Abstract
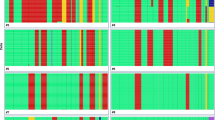
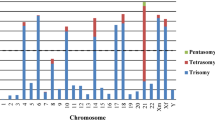
The detailed definition of karyotype changes associated with hyperdiploid acute lymphoblastic leukemia (ALL) is a precondition for their exploitation in minimal residual disease studies with fluorescence in situ hybridization analysis (FISH). In addition, certain karyotype patterns may have different prognostic implications. We have therefore used comparative genomic hybridization (CGH) to analyze the quantitative karyotype abnormalities in 14 cases of hyperdiploid ALL and correlated the results with those obtained by flow cytometry and conventional cytogenetic analyses. Despite an overall good agreement between the karyotypes obtained by classical banding techniques and CGH, we came across at least one karyotype discrepancy per case. Clarification of the discordant findings with fluorescence in situ hybridization (FISH) showed that all stem lines had been correctly defined by CGH. In eight cases, however, cytogenetic analyses revealed structural abnormalities that were undetectable by CGH. The other discrepancies were mainly due to a cytogenetic misinterpretation of similar sized and shaped chromosomes. Based on these findings we present a new diagnostic strategy for childhood ALL that includes flow cytometry and classical cytogenetics as well as CGH for the analysis of aneuploid cases and FISH to resolve the unavoidable discrepancies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Haas, O., Henn, T., Romanakis, K. et al. Comparative genomic hybridization as part of a new diagnostic strategy in childhood hyperdiploid acute lymphoblastic leukemia. Leukemia 12, 474–481 (1998). https://doi.org/10.1038/sj.leu.2400943
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.leu.2400943
Keywords
This article is cited by
-
Genome complexity in acute lymphoblastic leukemia is revealed by array-based comparative genomic hybridization
Oncogene (2007)
-
Identification of cryptic aberrations and characterization of translocation breakpoints using array CGH in high hyperdiploid childhood acute lymphoblastic leukemia
Leukemia (2006)