Abstract
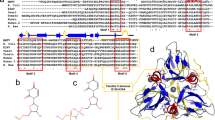
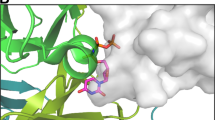
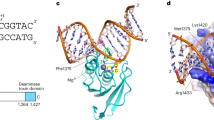
THE enzyme dUTPase catalyses the hydrolysis of dUTP1 and maintains a low intracellular concentration of dUTP so that uracil cannot be incorporated into DNA2. dUTPase from Escherichia coli is strictly specific for its dUTP substrate,3 the active site discriminating between nucleotides with respect to the sugar moiety as well as the pyrimidine base. Here we report the three-dimensional structure of E. coli dUTPase determined by X-ray crystallography at a resolution of 1.9Å. The enzyme is a symmetrical trimer, and of the 152 amino acid residues in the subunit, the first 136 are visible in the crystal structure. The tertiary structure resembles a jelly-roll fold and does not show the 'classical' nucleotide-binding domain. In the quaternary structure there is a complex interaction between the subunits that may be important in catalysis. This possibility is supported by the location of conserved elements in the sequence.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Komberg, A. & Baker, T. DNA Replication (Freeman, New York, 1991).
Tye, B. K. et al. Proc. natn. Acad. Sci. U.S.A. 74, 154–157 (1987).
Bertani, L. E., Häggmark, A. & Reichard, P. J. J. biol. Chem. 238, 3407–3413 (1963).
Hoffmann, I. et al. Eur. J. Biochem. 164, 45–51 (1987).
Cedergren-Zeppezauer, E. S. et al. Proteins: Struct. Funcn Genet. 4, 71–75 (1988).
Shlomai, J. & Komberg, A. J. biol. Chem. 253, 3305–3312 (1978).
Hokari, S. & Sakagishi, Y. Arch. biochem. Biophys. 253, 350–356 (1987).
Jones, E. Y., Stuart, D. I. & Walker, N. P. C. Nature 338, 225–228 (1989).
Eck, M. J. & Sprang, S. R. J. biol. Chem. 264, 17595–17605 (1989).
Branden, C.-I. & Tooze, J. in Introduction to Protein Structure 70–74 (Garland, New York, 1991).
Ladenstein, R. et al. J. molec. Biol. 203, 1045–1070 (1988).
Hoffmann, I. thesis. Univ. Saarbrücken, Germany (1988).
Lundberg, L. G., Thoresson, H.-O., Karlström, O. & Nyman, P. O. EMBO J. 2, 967–971 (1983).
McGeoch, D. J. Nucleic Acids Res. 18, 4105–4110 (1990).
Möller, W. & Amos, R. FEBS Lett. 186, 1–7 (1985).
Wang, B. C. Meth. Enzym. 115, 90–112 (1985).
Jones, T. A. J. appl. Crystallogr. 11, 268–272 (1978).
Brunger, A. T. J. molec. Biol. 203, 803–816 (1988).
Hendrickson, W. A. & Konnert, J. H. in Biomolecular Structure. Conformation. Function and Evolution Vol. 1 (ed. Srinivasan, R.) 43–57 (Pergamon, Oxford, 1980).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Cedergren-Zeppezauer, E., Larsson, G., Olof Nyman, P. et al. Crystal structure of a dUTPase. Nature 355, 740–743 (1992). https://doi.org/10.1038/355740a0
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/355740a0
This article is cited by
-
Redox status of cysteines does not alter functional properties of human dUTPase but the Y54C mutation involved in monogenic diabetes decreases protein stability
Scientific Reports (2021)
-
A High-Throughput Platform for the Generation of Synthetic Ab Clones by Single-Strand Site-Directed Mutagenesis
Molecular Biotechnology (2019)
-
Characterization of a dUTPase from the Hyperthermophilic Archaeon Thermococcus onnurineus NA1 and Its Application in Polymerase Chain Reaction Amplification
Marine Biotechnology (2007)
-
Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody
Nature (1998)
-
Crystal structure of a PDZ domain
Nature (1996)