Abstract
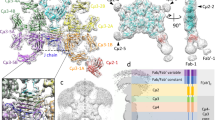
CRYSTAL structures of Fab antibody fragments determined by X-ray diffraction characteristically feature four-domain, β-barrel arrangements1–3. A human antibody Fc fragment has also been found to have four β-barrel domains4. The structures of a few intact antibodies have been solved5–8: in two myeloma proteins, the flexible hinge regions that connect the Fc to the Fab segments were deleted5,6 so the molecules were non-functional, structurally restrained, T-shaped antibodies; a third antibody, Kol, had no hinge residues missing but the Fc region was sufficiently disordered that it was not possible to relate its disposition accurately with respect to the Fab components7,8. Here we report the structure at 3.5 Å resolution of an IgG2a antitumour monoclonal antibody which contains an intact hinge region and was solved in a triclinic crystal by molecular replacement using known Fc and Fab fragments. The antibody is asymmetric, reflecting its dynamic character. There are two local, apparently independent, dyads in the molecule. One relates the heavy chains in the Fc, the other relates the constant domains of the Fabs. The variable domains are not related by this 2-fold axis because of the different Fab elbow angles of 159° and 143°. The Fc has assumed an asymmetric, oblique orientation with respect to loosely tethered yet almost collinear Fabs. Our study enables the two antigen-binding segments as well as the Fc portion of a functional molecule to be visualized and illustrates the flexibility of these immune response proteins.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Wilson, I. A., Rini, J. M., Fremont, D. H., Fieser, G. G. & Stura, E. A. Meth. Enzym. 203, 153–176 (1991).
Alzari, P. M., Lascombe, M.-B. & Poljak, R. J. A. Rev. Immun. 6, 555–580 (1988).
Davies, D. R., Padlan, E. A. & Sheriff, S. A. Rev. Biochem. 59, 439–473 (1990).
Deisenhofer, J. Biochemistry 20, 2361–2370 (1981).
Silverton, E. W., Navia, M. A. & Davies, D. R. Proc. natn. Acad. Sci. U.S.A. 74, 5140–5144 (1977).
Rajan, S. S. et al. Mol. Immun. 20, 797–799 (1983).
Colman, P. M., Deisenhofer, J., Huber, R. & Palm, W. J. molec. Biol. 100, 257–282 (1976).
Marquart, M., Deisenhofer, J., Huber, R. & Palm, W. J. molec. Biol. 141, 369–391 (1980).
Steplewski, Z., Jeglum, K. A., Rosales, C. & Weintraub, N. Cancer Immun. Immunother. 24, 197–201 (1987).
Rosales, C., Jeglum, K. A., Obrocka, M. & Steplewski, Z. Cell. Immun. 115, 420–428 (1988).
Jeglum, K. A. Proc. Seventh ACVIM Forum, San Diego (922–925) (Lippincott, Hagerstown, Maryland, 1989).
Larson, S., Day, J., Greenwood, A., Skaletsky, E. & McPherson, A. J. molec. Biol. 222, 17–19 (1991).
Fitzgerald, P. M. D. J. appl. Crystallogr. 21, 273–276 (1988).
Brunger, A. T., Kuriyan, J. & Karplus, M. Science 235, 458–460 (1987).
Brunger, A. T. A. Rev. phys. Chem. 42, 197–223 (1991).
Bernstein, F. C. et al. J. molec. Biol. 112, 535–542 (1977).
Crowther, R. A. in The Molecular Replacement Method (ed. Rossman, M. G.) 173–178 (Gordon and Breach, New York, 1972).
Sheriff, S. et al. Proc. natn. Acad. Sci. U.S.A. 84, 8075–8079 (1987).
Satow, Y., Cohen, G. H., Padlan, E. A. & Davies, D. R. J. molec. Biol. 190, 593–604 (1986).
Wrigley, N. G., Brown, E. B. & Skehel, J. J. J. molec. Biol. 169, 771–774 (1983).
Roux, K. H. Eur. J. Immun. 14, 459–464 (1984).
Lamy, J. et al. Biochemistry 24, 5532–5542 (1985).
Wade, R. H., Taveau, J. C. & Lamy, J. N. J. molec. Biol. 206, 349–356 (1989).
Schneider, W. P., Wensel, T. G., Stryer, L. & Oi, V. T. Proc. natn. Acad. Sci. U.S.A. 85, 2509–2513 (1988).
Dangl, J. L. et al. EMBO J. 7, 1989–1994 (1988).
Huber, R., Deisenhofer, J., Colman, P. M. & Matsushima, M. Nature 264, 415–420 (1976).
Amzel, L. M. & Poljak, R. J. A. Rev. Biochem. 48, 961–997 (1979).
Oi, V. T. et al. Nature 307, 136–140 (1984).
Jones, T. A. in Computational Crystallography (ed. Sayre, D.) 307–317 (Oxford University Press, New York, 1982).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Harris, L., Larson, S., Hasel, K. et al. The three-dimensional structure of an intact monoclonal antibody for canine lymphoma. Nature 360, 369–372 (1992). https://doi.org/10.1038/360369a0
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/360369a0
This article is cited by
-
Ligand functionalization of titanium nanopattern enables the analysis of cell–ligand interactions by super-resolution microscopy
Nature Protocols (2022)
-
Structural basis of polyethylene glycol recognition by antibody
Journal of Biomedical Science (2020)
-
Biophysical characterization of dynamic structures of immunoglobulin G
Biophysical Reviews (2020)
-
Conformation-controlled binding kinetics of antibodies
Scientific Reports (2016)
-
Immunoactive two-dimensional self-assembly of monoclonal antibodies in aqueous solution revealed by atomic force microscopy
Nature Materials (2014)