Abstract
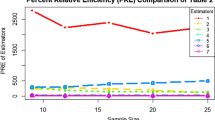
In order to evaluate the statistical properties of the variance components method implemented in SOLAR and GENEHUNTER2, the type I error rate, power and estimated size of modelled effects were determined using computer simulation. Results suggested that the type I error rate was quite conservative when the variance was completely due to random effects. However, when either a polygenic effect or an unlinked single locus component effect was included in the generating models the type I error rate was closer to the nominal level. The size of the polygenic or single locus effect appeared to influence the type I error rate. Results suggested that the variance components method underestimated the variance attributed to a single locus effect and overestimated the variance attributed to a polygenic effect in the models considered regardless of the source of variation.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Fisher RA . The correlation between relatives on the supposition of mendelian inheritance Roy Soc Edin 1918 52: 399–433
Penrose LS . Genetic linkage in graded human character Ann Eugenics 1937 8: 233–237
Jayakar SD . On the detection and estimation of linkage between a locus influencing a character and a marker locus Biometrics 1970 26: 451–464
Hopper JL, Mathews JD . Extensions to multivariate normal models for pedigree analysis Ann Hum Genet 1982 46: 373–383
Goldgar DE . Multipoint analysis for human quantitative genetic variation Am J Hum Genet 1990 47: 957–967
Schork NJ . Efficient computation of patterned covariance matrix mixed model in quantitative segregation analysis Genet Epidemiol 1991 8: 29–46
Schork NJ . Extended pedigree patterned covariance matrix mixed model for quantitative phenotype analysis Genet Epidemiol 1992 9: 73–86
Schork NJ . Extended multipoint identity-by-descent analysis of human quantitative traits: efficiency, power and modeling considerations Am J Hum Genet 1993 53: 1306–1319
Amos CI . Robust variance-components approach for assessing genetic linkage in pedigrees Am J Hum Genet 1994 54: 535–543
Fulker DW, Cherny SS . An improved multipoint sib-pair analysis of quantitative traits Behav Genet 1996 26: 527–532
Almasy L, Blangero J . Multipoint quantitative-trait linkage analysis in general pedigrees Am J Hum Genet 1998 62: 1198–1211
Sham PC, Cherny SS, Purcell S, Hewitt JK . Power of linkage versus association analysis of quantitative traits, by use of variance-components models, for sibship data Am J Hum Genet 2000 66: 1616–1630
Blangero J, Williams JT, Almasy L . Variance component methods for detecting complex trait loci Adv Genet 2001 42: 151–192
Pugh EW, Jaquish CE, Sorant AJ, Doestsch JP, Bailey-Wilson JE, Wilson AF . Comparison of sib-pair and variance components methods for genomic screening Genet Epidemiol 1997 14: 705–710
Shugart YY, Goldgar DE . Multipoint genomic scanning for quantitative loci: effects of map density, sibship size, and computational approach Eur J Hum Gene 1999 7: 103–109
William JT, Blangero J . Comparison of variance components and sibpair-based approaches to quantitative trait linkage analysis in unselected samples Genet Epidemiol 1999 16: 113–134
Wilson AF, Bailey-Wilson JE, Pugh EW, Sorant AJM . The genometric analysis simulation program (GASP): a software tool for testing and investigating methods in statistical genetics Am J Hum Genet 1996 59: A193
Allison DB, Neale MC, Zannolli R, Schork N, Amos CI, Blangero J . Testing the robustness of the likelihood-ratio test in a variance component quantitative trait loci mapping procedure Am J Hum Genet 1999 65: 531–544
Self SG, Liang KY . Asymptotic properties of maximum likelihood estimators likelihood ratio tests under non-standard conditions J Am Stat Assn 1987 82: 605–610
Lange KL, Little JA, Taylor JMG . Robust statistical modeling using the t distribution J Am Stat Assn 1989 84: 881–896
Astemborski JA, Beaty TH, Cohen BH . Variance components analysis on forced expiration in families Am J Hum Genet 1985 21: 741–753
Beaty TH, Liang KY . Robust inference for variance component models in families ascertained through probands. I. Conditional on proband's phenotype Genet Epidemiol 1987 4: 203–210
Beaty TH, Liang KY, Seerey S, Cohen BH . Robust inference for variance Genet Epidemiol 1987 4: 211–221
Beaty TH, Self SG, Liang KY, Connolly MA, Chase GA, Kwiterovich PO . Use of robust variance components method to analyze triglyceride data in families Ann Hum Genet 1985 49: 315–328
Acknowledgements
We thank three anonymous reviewers for insightful comments. This work was supported in part by Cigarette Restitution Fund of State of Maryland, NCI grant R03 CA85135-01 and NIH grant AG16992.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shugart, Y., O'Connell, J. & Wilson, A. An evaluation of the variance components approach: type I error, power and size of the estimated effect. Eur J Hum Genet 10, 133–136 (2002). https://doi.org/10.1038/sj.ejhg.5200772
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5200772
Keywords
This article is cited by
-
Heritability and Genetics of Serum Dickkopf 1 Levels in African Ancestry Families
Calcified Tissue International (2015)
-
Refining genome-wide linkage intervals using a meta-analysis of genome-wide association studies identifies loci influencing personality dimensions
European Journal of Human Genetics (2013)
-
Genome-wide linkage and peak-wide association study of obesity-related quantitative traits in Caribbean Hispanics
Human Genetics (2011)
-
The power to detect genetic linkage for quantitative traits in the Utah CEPH pedigrees
Journal of Human Genetics (2005)