Abstract
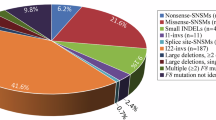
Phenylketonuric and hyperphenylalaninaemic patients in the population of the Republic of Ireland were screened for mutations at the human phenylalanine hydroxylase (PAH) locus. A composite data set for the island of Ireland was generated by merging the findings of this study with extant data for Northern Ireland. Analysis of this data on the basis of the four historic provinces (Munster, Leinster, Connacht and Ulster) revealed genetic diversity that is informative in terms of demographic forces that shaped the Irish population. R408W, the predominant Irish PAH mutation associated with haplotype 1.8, reached its highest relative frequency in the most westerly province, Connacht. This suggests that the gradient of R408W-1.8 observed across north-western Europe continues into Ireland and peaks in Connacht. Spatial autocorrelation analysis demonstrated that the gradient is consistent with a localised cline of R408W-1.8 likely to have been established by human migration. This and parallel allele frequency clines may represent the genetic traces of the Palaeolithic colonisation of Europe, a pattern not substantially altered in north-western Europe by subsequent Neolithic migrations. An analysis of mutant allele distributions in Ulster, Scotland and the rest of Ireland confirmed that Ulster has been a zone of considerable admixture between the Irish and Scottish populations, indicating a proportion of Scottish admixture in Ulster approaching 46%. Mutations primarily associated with Scandinavia accounted for 6.1% of mutations overall, illustrating the influence of Viking incursions on Irish population history.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Scriver CR . The Hyperphenylalaninaemias in Scriver CR, Beaudet AL, Sly WS, Valle D (eds) The Metabolic and Molecular Bases of Inherited Disease 7th edn New York: McGraw-Hill Inc. 1995 pp 1015–1075
Woo S, Lidsky A, Guttler F, Chandra T, Robson K . Cloned human phenylalanine hydroxylase gene permits prenatal and carrier detection of classical phenylketonuria Nature 1983 306: 151–155
Lidsky A, Robinson K, Thirumalachary C, Barker P, Ruddle F, Woo S . The PKU locus in man is on chromosome 12 Am J Hum Genet 1984 36: 527–533
Novacki P, Byck S, Prevost L, Scriver C . The PAH Mutation Analysis Consortium Database: Update 1996 Nucleic Acids Res 1997 25: 139–142
Scriver CR, Waters PJ, Sarkissian C et al. PAHdb: A locus-specific knowledgebase Hum Mutat 2000 15: 99–104
DiLella A, Kwok S, Ledley F, Marvit J, Woo S . Molecular structure and polymorphic map of the human phenylalanine hydroxylase gene Biochemistry 1986 25: 749–753
Woo S . Collation of RFLP haplotypes at the human phenylalaninehydroxylase (PAH) locus Am J Hum Genet 1988 43: 781–783
Goltsov AA, Eisensmith RC, Konecki DS, Lichter-Konecki U, Woo SL . Associations between mutations and a VNTR in the human phenylalanine hydroxylase gene Am J Hum Genet 1992 51: 627–636
Goltsov A, Eisensmith R, Naughten E, Jin L, Chakraborty R, Woo S . A single polymorphic STR system in the human phenylalanine hydroxylase gene permits rapid prenatal diagnosis and carrier screening for phenylketonuria Hum Molec Genet 1993 2: 577–581
Chakraborti R, Lidsky A, Daiger S et al. Polymorphic DNA haplotypes at the human phenylalanine hydroxylase locus and their relationship with phenylketonuria Hum Genet 1987 76: 40–46
Daiger S, Chakraborty R, Reed L et al. Polymorphic DNA haplotypes at the phenylalanine hydroxylase (PAH) locus in European families with phenylketonuria (PKU) Am J Hum Genet 1989 45: 310–318
Sullivan SE, Moore SD, Connor JM et al. Haplotype distribution of the human phenylalanine hydroxylase locus in Scotland and Switzerland Am J Hum Genet 1989 44: 652–659
Konecki D, Lichter-Konecki U . The phenylalanine hydroxylase locus: current knowledge about alleles and mutations of the phenylalanine hydroxylase gene in various populations Hum Genet 1991 87: 377–388
Eisensmith R, Woo S . Updated listing of haplotypes at the human phenylalanine hydroxylase (PAH) locus Am J Hum Genet 1992 51: 1445–1448
Carter KC, Byck S, Waters PJ et al. Mutation at the phenylalanine hydroxylase gene (PAH) and its use to document population genetic variation: the Quebec experience Eur J Hum Genet 1998 6: 61–70
O'Neill C, Eisensmith RC, Croke D, Naughten ER, Cahalane SF, Woo SLC . Molecular analysis of PKU in Ireland Acta Paediatrica 1995 407: 43–44
Zschocke J, Graham C, Carson DJ, Nevin N . Phenylketonuria mutation analysis in Northern Ireland: a rapid stepwise approach Am J Hum Genet 1995 57: 1311–1317
Zschocke J, Mallory JP, Eiken HG, Nevin N . Phenylketonuria and the peoples of Northern Ireland Hum Genet 1997 100: 189–194
Eisensmith RC, Goltsov AA, O'Neill C et al. Recurrence of the R408W mutation in the Phenylalanine Hydroxylase locus in Europeans Am J Hum Genet 1995 56: 278–286
Guthrie R, Susi A . A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants Pediatrics 1963 32: 338–343
Guldberg P, Guttler F . ‘Broad range’ DGGE for single-step mutation scanning of entire genes: application to human phenylalanine hydroxylase gene Nucleic Acids Res 1994 22: 880–881
Rao V . Direct sequencing of polymerase chain reaction amplified DNA Anal Biochem 1994 216: 1–14
Bodmer W, Cavalli-Sforza LL . The genetics of human populations San Francisco, USA: WH Freeman & Company 1971
Sokal RR, Oden NL . Spatial autocorrelation analysis in biology Biol J Linn Soc 1978 10: 199–249
Sokal RR . Ecological parameters inferred from spatial correlograms in Patil GP, Rosenzweig M (eds) Contemporary Quantitative Ecology and Related Ecometrics Fairland, MD, USA: International Co-operative Publishing House 1979 pp 167–196
Oden NL . Assessing the significance of a spatial correlogram Geogr Anal 1984 16: 1–16
Bernstein F . Die geographische verteilung der blut-gruppen und ihre anthropologische bedeutung in: Comitato Italiano per o studio dei problemi della populazione Roma, Italia: Instituto Poligrafico dello Stato 1931 pp 227–243
Bertorelle G, Excoffier L . Inferring admixture proportions from molecular data Mol Biol Evol 1998 15: 1298–1311
Dworniczak B, Wedemeyer N, Horst J . PCR detection of the BglII RFLP at the human phenylalanine hydroxylase (PAH) locus Nucleic Acids Res 1991 19: 1958
Iyengar S, Seaman M, Deinard AS et al. Analyses of cross-species polymerase chain reaction products to infer the ancestral state of human polymorphisms DNA Sequence 1998 8: 317–327
Wedemeyer N, Dworniczak B, Horst J . PCR detection of the MspI (Aa) RFLP at the human phenylalanine hydroxylase (PAH) locus Nucleic Acids Res 1991 19: 1959
Goltsov AA, Woo SL . Detection of the XmnI RFLP at the human phenylalanine hydroxylase locus by PCR Genbank accession number Z11537 1992
Zschocke J, Graham C, McKnight J, Nevin N . The STR system in the human phenylalanine hydroxylase gene: true fragment length obtained with fluorescent labelled PCR primers Acta Paed Suppl 1994 407: 41–42
Tyfield L, Stephenson A, Cockburn F et al. Sequence variation at the phenylalanine hydroxylase gene in the British Isles Am J Hum Genet 1997 60: 388–396
Eiken HG, Knappskog PM, Boman H et al. Relative frequency, heterogeneity and geographic clustering of PKU mutations in Norway Eur J Hum Genet 1996 4: 205–213
Mallolas J, Vilaseca MA, Campistol J et al. Mutational spectrum of phenylalanine hydroxylase deficiency in the population resident in Catalonia: genotype-phenotype correlation Hum Genet 1999 105: 468–473
Dworniczak B, Kalaydjieva L, Pankoke S, Aulehla-Scholz C, Allen G, Horst J . Analysis of exon 7 of the human phenylalanine hydroxylase gene: a mutation hot spot? Hum Mutat 1992 1: 138–146
Guldberg P, Levy H, Handley W et al. Phenylalanine hydroxylase gene mutations in the United States: Report from the Maternal PKU collaborative study Am J Hum Genet 1996 59: 84–94
Byck S, Morgan K, Tyfield L, Dworniczak B, Scriver CR . Evidence for origin, by recurrent mutation, of the phenylalanine hydroxylase R408W mutation on two haplotypes in European and Quebec populations Hum Molec Genet 1994 3: 1675–1677
Charles-Edwards TM . Early Christian Ireland Cambridge, UK: Cambridge University Press 2000
Relethford JH, Lees FC, Crawford MH . Population structure and anthropometric variation in rural Western Ireland: migration and biological differentiation Ann Hum Biol 1980 7: 411–428
Kalaydjieva L, Dworniczak B, Kucinskas V, Yurgeliavicius V, Kunert E, Horst J . Geographical distribution gradients of the major PKU mutations and the linked haplotypes Hum Genet 1991 86: 411–413
Giannattasio S, Jurgelevicius V, Lattanzio P, Cimbalistiene L, Marra E, Kucinskas V . Phenylketonuria mutations and linked haplotypes in the Lithuanian population: origin of the most common R408W mutation Hum Hered 1997 47: 155–160
Treacy E, Byck S, Clow C, Scriver C . ‘Celtic’ phenylketonuria chromosomes found? Evidence in two regions of Quebec Province Eur J Hum Genet 1993 1: 220–228
Treacy E . Phenylketonuria, a Celtic condition revisited Ir Med J 1994 87: 100
Murphy M, McHugh B, Tighe O et al. The genetic basis of transferase-deficient galactosaemia in Ireland and the population history of the Irish Travellers Eur J Hum Genet 1999 7: 549–554
O Corrain D . Ireland, Wales, Man and the Hebrides in Sawyer P. (ed) The Oxford Illustrated History of the Vikings Oxford: Oxford University Press 1997 pp 83–109
Helgason A, Sigurdardottir S, Gulcher J, Ward R, Stefansson K . mtDNA and the origin of the Icelanders: deciphering signals of recent population history Am J Hum Genet 2000 66: 999–1016
Helgason A, Sigurdardottir S, Nicholson J et al. Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland Am J Hum Genet 2000 67: 697–717
Helgason A, Hickey E, Goodacre S et al. mtDNA and the islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry Am J Hum Genet 2001 68: 723–737
Dawson GWP . The frequencies of the ABO and Rh (D) blood groups in Ireland from a sample of 1 in 18 of the population Ann Hum Genet 1964 28: 49–59
Hill EW, Jobling MA, Bradley DJ . Y-chromosome variation and Irish origins Nature 2000 404: 351–352
Cavalli-Sforza LL, Menozzi P, Piazza A . The History and Geography of Human Genes Princeton, NJ: Princeton University Press 1994
Barbujani G, Bertorelle G . Genetics and the population history of Europe Proc Natl Acad Sci USA 2001 98: 22–25
Flanagan MT . The Vikings in Loughrey P (ed) The people of Ireland Belfast, Northern Ireland: Appletree 1988 pp 55–84
Mallory JP, McNeill TE . The Archaeology of Ulster Belfast, N. Ireland: Institute of Irish Studies 1989
Apold J, Eiken H, Odland E et al. A termination mutation prevalent in Norwegian haplotype 7 phenylketonuria genes Am J Hum Genet 1990 47: 1002–1007
Acknowledgements
We thank our colleagues Dr Donncha Dunican, Dr Peter McWilliam, Professor Stanley Monkhouse, Dr Daniel Bradley (Smurfit Institute of Genetics, Trinity College Dublin), Dr Barra O Donnabhain (Department of Archaeology, University College Cork) and Dr Linda Tyfield (University of Bristol, UK) for helpful discussions and for critical reading of the manuscript. We gratefully acknowledge financial support from the Research Committee of the Royal College of Surgeons in Ireland and from the Royal Irish Academy Millennium Project on ‘Irish Origins: the genetic history and geography of Ireland’ jointly funded with the National Millennium Committee of Ireland.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Assume that the overall mutant allele frequencies q1 and q2 can be estimated in the parental populations 1 and 2, for example from the square root of the disease incidence in these populations assuming Hardy–Weinberg equilibrium. The incidence of the disease in the hybrid group, however, is not known and the overall mutant allele frequency qh cannot be estimated. Assume also that the relative frequency of a specific mutant, ie the frequency of that allele within a sample of mutants, is known for the parental populations (r1 and r2) and for the hybrid population (rh). The Berstein model27 suggests that the frequency of an allele in the hybrid population (rhqh in our case) is a linear combination of the frequencies of the allele in the parental populations (r1q1 and r2q2), that is:

where m and (1-m) are the relative contributions of parental population 1 and parental population 2 into the hybrid respectively. In our case, however, qh is unknown and m thus cannot be estimated directly from equation 1. A possible solution is to assume that the incidence of the disease in the hybrid group, and therefore qh, is also a linear combination of the incidences observed in the parental populations. That is:

Combining equations (1) and (2) we obtain the following estimate of m:

As suggested for other estimators of a ratio28, we propose to combine the information from different mutants averaging first the numerator and the denominator of (3) across mutations, and taking then the ratio of these averages. The estimate of m can be finally used to compute back an estimate of qh from equation (2).
Rights and permissions
About this article
Cite this article
O'Donnell, K., O'Neill, C., Tighe, O. et al. The mutation spectrum of hyperphenylalaninaemia in the Republic of Ireland: the population history of the Irish revisited. Eur J Hum Genet 10, 530–538 (2002). https://doi.org/10.1038/sj.ejhg.5200841
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5200841
Keywords
This article is cited by
-
Genotype–phenotype correlations analysis of mutations in the phenylalanine hydroxylase (PAH) gene
Journal of Human Genetics (2008)