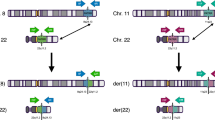
Abstract
Approximately one in 500 individuals carries a reciprocal translocation. Of the 121 monosomy 1p36 subjects ascertained by our laboratory, three independent cases involved unbalanced translocations of 1p and 9q, all of which were designated t(1;9)(p36.3;q34). These derivative chromosomes were inherited from balanced translocation carrier parents. To understand better the causes and consequences of chromosome breakage and rearrangement in the human genome, we characterized each derivative chromosome at the DNA sequence level and identified the junctions between 1p36 and 9q34. The breakpoint regions were unique in all individuals. Insertions and duplications were identified in two balanced translocation carrier parents and their unbalanced offspring. Sequence analyses revealed that the translocation breakpoints disrupted genes. This study demonstrates that apparently balanced reciprocal translocations in phenotypically normal carriers may have cryptic imbalance at the breakpoints. Because disrupted genes were identified in the phenotypically normal translocation carriers, caution should be exercised when interpreting data on phenotypically abnormal carriers with apparently balanced rearrangements that disrupt putative candidate genes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Mansouri MR, Carlsson B, Davey E et al: Molecular genetic analysis of a de novo balanced translocation t(6;17)(p21.31;q11.2) associated with hypospadias and anorectal malformation. Hum Genet 2006; 119: 162–168.
Bodrug SE, Holden JJ, Ray PN, Worton RG : Molecular analysis of X-autosome translocations in females with Duchenne muscular dystrophy. EMBO J 1991; 10: 3931–3939.
Bodrug SE, Ray PN, Gonzalez IL, Schmickel RD, Sylvester JE, Worton RG : Molecular analysis of a constitutional X-autosome translocation in a female with muscular dystrophy. Science 1987; 237: 1620–1624.
Haider S, Matsumoto R, Kurosawa N, Wakui K, Fukushima Y, Isobe M : Molecular characterization of a novel translocation t(5;14)(q21;q32) in a patient with congenital abnormalities. J Hum Genet 2006; 51: 335–340.
McMullan TW, Crolla JA, Gregory SG et al: A candidate gene for congenital bilateral isolated ptosis identified by molecular analysis of a de novo balanced translocation. Hum Genet 2002; 110: 244–250.
Millar JK, Wilson-Annan JC, Anderson S et al: Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 2000; 9: 1415–1423.
Schule B, Albalwi M, Northrop E et al: Molecular breakpoint cloning and gene expression studies of a novel translocation t(4;15)(q27;q11.2) associated with Prader-Willi syndrome. BMC Med Genet 2005; 6: 18.
Yoshiura K, Machida J, Daack-Hirsch S et al: Characterization of a novel gene disrupted by a balanced chromosomal translocation t(2;19)(q11.2;q13.3) in a family with cleft lip and palate. Genomics 1998; 54: 231–240.
Ballif BC, Kashork CD, Shaffer LG : FISHing for mechanisms of cytogenetically defined terminal deletions using chromosome-specific subtelomeric probes. Eur J Hum Genet 2000; 8: 764–770.
Heilstedt HA, Ballif BC, Howard LA et al: Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet 2003; 72: 1200–1212.
Gajecka M, Pavlicek A, Glotzbach CD et al: Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations. Hum Genet 2006, (in press).
Shapira SK, McCaskill C, Northrup H et al: Chromosome 1p36 deletions: the clinical phenotype and molecular characterization of a common newly delineated syndrome. Am J Hum Genet 1997; 61: 642–650.
Wu YQ, Heilstedt HA, Bedell JA et al: Molecular refinement of the 1p36 deletion syndrome reveals size diversity and a preponderance of maternally derived deletions. Hum Mol Genet 1999; 8: 313–321.
Yu W, Ballif BC, Kashork CD et al: Development of a comparative genomic hybridization microarray and demonstration of its utility with 25 well-characterized 1p36 deletions. Hum Mol Genet 2003; 12: 2145–2152.
Shaffer LG, McCaskill C, Han JY et al: Molecular characterization of de novo secondary trisomy 13. Am J Hum Genet 1994; 55: 968–974.
Rautenstrauss B, Fuchs C, Liehr T, Grehl H, Murakami T, Lupski JR : Visualization of the CMT1A duplication and HNPP deletion by FISH on stretched chromosome fibers. J Peripher Nerv Syst 1997; 2: 319–322.
Page SL, Shaffer LG : Nonhomologous Robertsonian translocations form predominantly during female meiosis. Nat Genet 1997; 15: 231–232.
Ballif BC, Wakui K, Gajecka M, Shaffer LG : Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Hum Genet 2004; 114: 198–206.
Kent WJ : BLAT – the BLAST-like alignment tool. Genome Res 2002; 12: 656–664.
Karolchik D, Baertsch R, Diekhans M et al: The UCSC Genome Browser Database. Nucleic Acids Res 2003; 31: 51–54.
Jurka J : Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 2000; 16: 418–420.
Benson G : Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 1999; 27: 573–580.
Warburton D : De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. Am J Hum Genet 1991; 49: 995–1013.
Wirth J, Nothwang HG, van der Maarel S et al: Systematic characterisation of disease associated balanced chromosome rearrangements by FISH: cytogenetically and genetically anchored YACs identify microdeletions and candidate regions for mental retardation genes. J Med Genet 1999; 36: 271–278.
Astbury C, Christ LA, Aughton DJ et al: Detection of deletions in de novo ‘balanced’ chromosome rearrangements: further evidence for their role in phenotypic abnormalities. Genet Med 2004; 6: 81–89.
Gribble SM, Prigmore E, Burford DC et al: The complex nature of constitutional de novo apparently balanced translocations in patients presenting with abnormal phenotypes. J Med Genet 2005; 42: 8–16.
Baptista J, Prigmore E, Gribble SM, Jacobs PA, Carter NP, Crolla JA : Molecular cytogenetic analyses of breakpoints in apparently balanced reciprocal translocations carried by phenotypically normal individuals. Eur J Hum Genet 2005; 13: 1205–1212.
Acknowledgements
We thank Aaron Theisen (Washington State University, Spokane, WA) for his critical editing of the manuscript and Adam Pavlicek (Genetic Information Research Institute, Mountain View, CA, USA) for examining the breakpoint junctions of the subjects.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gajecka, M., Glotzbach, C., Jarmuz, M. et al. Identification of cryptic imbalance in phenotypically normal and abnormal translocation carriers. Eur J Hum Genet 14, 1255–1262 (2006). https://doi.org/10.1038/sj.ejhg.5201710
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201710
Keywords
This article is cited by
-
Unrevealed mosaicism in the next-generation sequencing era
Molecular Genetics and Genomics (2016)
-
Recurrence risks for different pregnancy outcomes and meiotic segregation analysis of spermatozoa in carriers of t(1;11)(p36.22;q12.2)
Journal of Human Genetics (2014)
-
Copy number variants and infantile spasms: evidence for abnormalities in ventral forebrain development and pathways of synaptic function
European Journal of Human Genetics (2011)
-
Further delineation of nonhomologous-based recombination and evidence for subtelomeric segmental duplications in 1p36 rearrangements
Human Genetics (2009)
-
Identification of sequence motifs at the breakpoint junctions in three t(1;9)(p36.3;q34) and delineation of mechanisms involved in generating balanced translocations
Human Genetics (2006)