Abstract
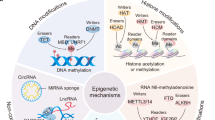
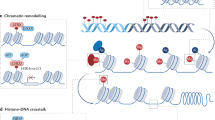
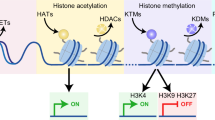
Significant evidences have brought new insights on the mechanisms by which epigenetic machinery proteins regulate gene expression, leading to a redefinition of chromatin regulation in terms of modification of core histones, DNA methylation, RNA-mediated silencing pathways, action of methylation-dependent sensitive insulators and Polycomb/Trithorax group proteins. Consistent with these fundamental aspects, an increasing number of human pathologies have been found to be associated with aberrant epigenetics regulation, including cancer, mental retardation, neurodegenerative symptoms, imprinting disorders, syndromes involving chromosomal instabilities and a great number of human life-threatening diseases. The possibility of reversing epigenetic marks, in contrast to genetic code, may provide new pharmacological targets for emerging therapeutic intervention.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ausió J, Levin DB, de Amorim GV, Bakker S, Macleod PM : Syndromes of disordered chromatin remodeling. Clin Genet 2003; 64: 83–95.
Thomson S, Mahadevan LC, Clayton AL : MAP kinase-mediated signaling to nucleosomes and immediate-early gene induction. Semin Cell Dev Biol 1999; 10: 205–214.
Downs JA, Lowndens NF, Jackson SP : A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 2000; 408: 1001–1004.
Jacquot S, Zeniou M, Touraine R, Hanauer A : X-linked Coffin–Lowry syndrome (CLS, MIM 303600, RPS6KA3 gene, protein product known under various names: pp. 90 (rsk2), RSK2, ISPK, MAPKAP1). Eur J Hum Genet 2002; 10: 2–5.
Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL : Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell 2001; 107: 727–738.
Plath K, Fang J, Mlynarczyk-Evans SK et al: Role of histone H3 lysine 27 methylation in X inactivation. Science 2003; 300: 131–135.
Rice JC, Briggs SD, Ueberheide B et al: Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 2003; 12: 1591–1598.
Bannister AJ, Zegerman P, Partridge JF et al: Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 2001; 410: 120–124.
Sun XJ, Wei J, Wu XY et al: Identification and characterization of a novel human histone H3 lysine 36-specific methyltransferase. J Biol Chem 2005; 280: 35261–35271.
Roberts CW, Orkin SH : The SWI/SNF complex – chromatin and cancer. Nat Rev Cancer 2004; 4: 133–142.
Picketts DJ, Higgs DR, Bachoo S, Blake DJ, Quarrell OW, Gibbons RJ : ATRX encodes a novel member of the SNF2 family of proteins: mutations point to a common mechanism underlying the ATR-X syndrome. Hum Mol Genet 1996; 5: 1899–1907.
Bartolomei MS, Tilghman SM : Genomic imprinting in mammals. Annu Rev Genet 1997; 31: 493–525.
Li E, Chen T, Dodge J, Hata K, Okano M, Ueda Y : DNA methylation in development and diseases. Human Genome Meeting Symposium Abstracts: 2003, Presentation 13. 2003, http://hgm2003.hgu.mrc.ac.uk/Abstracts/Publish/Symposia/Symposium04/hgm13.html.
Hornstra I K, Yang TP : High resolution methylation analysis of the human HPRT gene 5′ region on the active and inactive X chromosomes: correlation with binding sites for transcriptional factors. Mol Cell Biol 1994; 14: 1419–1430.
Jones PA : DNA methylation errors and cancer. Cancer Res 1996; 56: 2463–2467.
Cleveland DW, Mao Y, Sullivan KF : Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 2003; 112: 407–421.
Richardson B : DNA methylation and autoimmune disease. Clin Immunol 2003; 109: 72–79.
Issa JP : CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol 2000; 249: 101–118.
Levenson JM, Roth TL, Lubin FD et al: Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem 2006; 281: 15763–15773.
James SJ, Pogribna M, Pogribny IP et al: Abnormal folate metabolism and mutation in the methylenetetrahydrofolate reductase gene may be maternal risk factors for Down syndrome. Am J Clin Nutr 1999; 70: 495–501.
van der Put NM, Thomas CM, Eskes TK et al: Altered folate and vitamin B12 metabolism in families with spina bifida offspring. Q J Med 1997; 90: 505–510.
Murray JC : Gene/environment causes of cleft lip and/or palate. Clin Genet 2002; 61: 248–256.
Hassold TJ, Burrage LC, Chan ER et al: Maternal folate polymorphisms and the etiology of human nondisjunction. Am J Hum Genet 2001; 69: 434–439.
Skibola CF, Smith MT, Kane E et al: Polymorphisms in the methylenetetrahydrofolate reductase gene are associated with susceptibility to acute leukemia in adults. Proc Natl Acad Sci USA 1999; 96: 12810–12815.
Boushey CJ, Beresford SA, Omenn GS, Motulsky AG : A quantitative assessment of plasma homocysteine as a risk factor for vascular disease. Probable benefits of increasing folic acid intakes. JAMA 1995; 274: 1049–1057.
Hankey GJ, Eikelboom JW : Homocysteine and stroke. Curr Opin Neurol 2001; 14: 95–102.
Sun J, Xu Y, Zhu Y, Lu H : Genetic polymorphism of methylenetetrahydrofolate reductase as a risk factor for diabetic nephropathy in Chinese type 2 diabetic patients. Diabetes Res Clin Pract 2004; 64: 185–190.
Herran A, Garcia-Unzueta MT, Amado JA, Lopez-Cordovilla JJ, Diez-Manrique JF, Vazquez-Barquero JL : Folate levels in psychiatric outpatients. Psychiatry Clin Neurosci 1999; 53: 531–533.
Mattson MP : Methylation and acetylation in nervous system development and neurodegenerative disorders. Ageing Res Rev 2003; 2: 329–342.
Miller A : The methionine-homocysteine cycle and its effect on cognitive diseases. Alt Med Rev 2003; 8: 7–19.
Mathe G : Why have ten or so nontoxic, retrovirus integrase inhibitors not been made available for AIDS treatment? A ten-year experience [correction of experiment] must liberate them. Biomed Pharmacother 1999; 53: 484–486.
Lucock M, Yates Z : Folic acid – vitamin and panacea or genetic time bomb? Nat Rev Genet 2005; 6: 235–240.
Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T : DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet 2000; 24: 88–91.
Jeltsch A, Nellen W, Lyko F : Two substrates are better than one: dual specificities for Dnmt2 methyltransferases. Trends Biochem Sci 2006; 31: 306–308.
Okano M, Bell DW, Haber DA, Li E : DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99: 247–257.
Kareta MS, Botello ZM, Ennis JJ, Chou C, Chedin F : Reconstitution and mechanism of the stimulation of DE NOVO methylation by human DNMT3L. J Biol Chem 2006; July 7; [Epub ahead of print].
Robertson KD, Uzvolgyi E, Liang G et al: The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res 1999; 27: 2291–2298.
Chen CL, Yan X, Gao YN, Liao QP : Expression of DNA methyltransferase 1, 3A and 3B mRNA in the epithelial ovarian carcinoma. Zhonghua Fu Chan Ke Za Zhi 2005; 40: 770–774.
Karpf AR, Matsui S : Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res 2005; 65: 8635–8639.
Xu GL, Bestor TH, Bourc'his D et al: Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 1999; 402: 187–191.
Jones PL, Veenstra GJ, Wade PA et al: Methylated DNA and MECP2 recruit histone deacetylase to repress transcription. Nat Genet 1998; 19: 187–191.
Moog U, Smeets EE, Roozendaal KEV et al: Neurodevelopmental disorders in males related to the gene causing Rett syndrome in females (MECP2). Eur J Paediatr Neurol 2003; 7: 5–12.
Collins AL, Levenson JM, Vilaythong AP et al: Mild overexpression of MeCP2 causes a progressive neurological disorder in mice. Hum Mol Genet 2004; 13: 2679–2689.
Zhu Y, Spitz MR, Zhang H, Grossman HB, Frazier ML, Wu X : Methyl-CpG-binding domain 2: a protective role in bladder carcinoma. Cancer 2004; 100: 1853–1858.
Bader S, Walker M, McQueen HA et al: MBD1, MBD2 and CGBP genes at chromosome 18q21 are infrequently mutated in human colon and lung cancers. Oncogene 2003; 22: 3506–3510.
O'Donnell WT, Warren ST : A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci 2002; 25: 315–338.
Doerfler W : Patterns of DNA methylation – evolutionary vestiges of foreign DNA inactivation as a host defense mechanism. A proposal. Biol Chem Hoppe Seyler 1991; 372: 557–564.
Kafri T, Ariel M, Brandeis M et al: Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev 1992; 6: 705–714.
Rakyan V, Whitelaw E : Transgenerational epigenetic inheritance. Curr Biol 2003; 13: R6.
Moore T, Haig D : Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 1991; 7: 45–49.
Nicholls RD, Knepper JL : Genome organization, function, and imprinting in Prader–Willi and Angelman syndromes. Annu Rev Genomics Hum Genet 2001; 2: 153–175.
Maison C, Bailly D, Peters AH et al: High-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet 2002; 30: 329–334.
Avner P, Heard E : X-chromosome inactivation: counting, choice and initiation. Nat Rev Genet 2001; 2: 59–67.
Schramke V, Allshire R : Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 2003; 301: 1069–1073.
Calin GA, Sevignani C, Dumitru CD et al: Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancer. Proc Natl Acad Sci 2004; 101: 2999–3004.
Calin GA, Dumitru CD, Shimizu M et al: Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci 2002; 99: 15524–15529.
Sleutels F, Zwart R, Barlow DP : The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 2002; 415: 810–813.
Tufarelli C, Stanley JA, Garrick D et al: Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 2003; 34: 157–165.
Huang L, Gledhill J, Cameron CE : RNA-dependent RNA polymerase in gene silencing; In Hannon G (ed): RNAi: a guide for gene silencing. Cold Spring Habor Laboratory Press: NY, 2003, pp 175–203.
Verdel A, Jia S, Gerber S et al: RNAi-mediated targeting of heterochromatin by the RITS complex. Science 2004; 303: 672–676.
Noma K, Sugiyama T, Cam H et al: RITS acts in cis to promote RNA interference-mediated transcriptional and posttranscriptional silencing. Nat Genet 2004; 36: 1174–1180.
Kawasaki H, Taira K : Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 2004; 431: 211–217.
Robertson KD : DNA methylation and human disease. Nat Rev Genet 2005; 6: 597–610.
Arney KL : H19 and Igf2 – enhancing the confusion? Trends Genet 2003; 19: 17–23.
Kono T, Obata Y, Wu Q et al: Birth of parthenogenetic mice that can develop to adulthood. Nature 2004; 428: 860–864.
Orlando V : Polycomb, epigenomes and control of cell identity. Cell 2003; 112: 599–606.
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D : Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 2002; 16: 2893–2905.
Nakamura T, Mori T, Tada S et al: ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 2002; 10: 1119–1128.
Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B : Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 2002; 36: 233–278.
Johnston CM, Newall AE, Brockdorff N, Nesterova TB : Enox, a novel gene that maps 10 kb upstream of Xist and partially escapes X inactivation. Genomics 2002; 80: 236–244.
Ogawa Y, Lee JT : Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol Cell 2003; 11: 731–743.
O'Neill LP, Keohane AM, Lavender JS et al: A developmental switch in H4 acetylation upstream of Xist plays a role in X chromosome inactivation. EMBO J 1999; 18: 2897–2907.
Chao W, Huynh KD, Spencer RJ, Davidow LS, Lee JT : CTCF, a candidate trans-acting factor for X-inactivation choice. Science 2002; 295: 345–347.
Silva J, Mak W, Zvetkova I et al: Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell 2003; 4: 481–495.
Varambally S, Dhanasekaran SM, Zhou M et al: The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002; 419: 624–629.
Raaphorst FM, Meijer CJ, Fieret E et al: Poorly differentiated breast carcinoma is associated with increased expression of the human polycomb group EZH2 gene. Neoplasia 2003; 5: 481–488.
Gabellini D, Green MR, Tupler R : Inappropriate gene activation in FSHD: a repressor complex bnds a chromosomal repeat deleted in dystrophic muscle. Cell 2002; 110: 339–348.
Saunthararajah Y, Hillery CA, Lavelle D et al: Effects of 5-aza-2′-deoxycytidine on fetal hemoglobin levels, red cell adhesion, and hematopoietic differentiation in patients with sickle cell disease. Blood 2003; 102: 3865–3870.
Issa JP, Garcia-Manero G, Giles FJ et al: Phase I study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 2004; 103: 1635–1640.
Mizugaki M, Yamaguchi T, Ishiwata S et al: Alteration of DNA methylation levels in MRL lupus mice. Clin Exp Immunol 1997; 110: 265–269.
Tabolacci E, Pietrobono R, Moscato U, Oostra BA, Chiurazzi P, Neri G : Differential epigenetic modifications in the FMR1 gene of the fragile X syndrome after reactivating pharmacological treatments. Eur J Hum Genet 2005; 13: 641–648.
Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE : Specific inhibition of DNMT1 by antisense oligonucleotides induces re-expression of estrogen receptor-alpha (ER) in ER-negative human breast cancer cell lines. Cancer Biol Ther 2003; 2: 552–556.
Marks PA, Miller T, Richon VM : Histone deacetylases. Curr Opin Pharmacol 2003; 3: 344–351.
Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS : Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem 2001; 276: 36734–36741.
Komatsu N, Kawamata N, Takeuchi S et al: SAHA, a HDAC inhibitor, has profound anti-growth activity against non-small cell lung cancer cells. Oncol Rep 2006; 15: 187–191.
Marks PA, Dokmanovic M : Histone deacetylase inhibitors: discovery and development as anticancer agents. Expert Opin Investig Drugs 2005; 14: 1497–1511.
Kee HJ, Sohn IS, Nam KI et al: Inhibition of histone deacetylation blocks cardiac hypertrophy induced by angiotensin II infusion and aortic banding. Circulation 2006; 113: 51–59.
Camelo S, Iglesias AH, Hwang D et al: Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol 2005; 164: 10–21.
McCampbell A, Taye AA, Whitty L, Penney E, Steffan JS, Fischbeck KH : Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc Natl Acad Sci USA 2001; 98: 15179–15184.
Hockly E, Richon VM, Woodman B et al: Suberoylanilidehydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc Natl Acad Sci USA 2003; 100: 2041–2046.
Minamiyama M, Katsuno M, Adachi H et al: Sodium butyrate ameliorates phenotypic expression in a transgenic mouse model of spinal and bulbar muscular atrophy. Hum Mol Genet 2004; 13: 1183–1192.
Belinsky SA, Klinge DM, Stidley CA et al: Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res 2003; 63: 7089–7093.
Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R : Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res 2000; 60: 6039–6044.
Chinnaiyan P, Vallabhaneni G, Armstrong E, Huang SM, Harari PM : Modulation of radiation response by histone deacetylase inhibition. Int J Radiat Oncol Biol Phys 2005; 62: 223–229.
Karpf AR, Jones DA : Reactivating the expression of methylation silenced genes in human cancer. Oncogene 2002; 21: 5496–5503.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Santos-Rebouças, C., Pimentel, M. Implication of abnormal epigenetic patterns for human diseases. Eur J Hum Genet 15, 10–17 (2007). https://doi.org/10.1038/sj.ejhg.5201727
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/sj.ejhg.5201727
Keywords
This article is cited by
-
The effects of exercise on epigenetic modifications: focus on DNA methylation, histone modifications and non-coding RNAs
Human Cell (2024)
-
Cognitive Dysfunction and Exercise: From Epigenetic to Genetic Molecular Mechanisms
Molecular Neurobiology (2024)
-
Particulate matter exposure shapes DNA methylation through the lifespan
Clinical Epigenetics (2019)
-
Investigating the genetic and epigenetic basis of big biological questions with the parthenogenetic marbled crayfish: A review and perspectives
Journal of Biosciences (2018)
-
Physical activity in the prevention of human diseases: role of epigenetic modifications
BMC Genomics (2017)