ABSTRACT
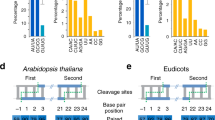
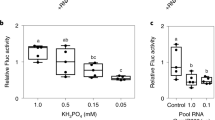
Seventy-five previously known plant microRNAs (miRNAs) were classified into 14 families according to their gene sequence identity. A total of 18,694 plant expressed sequence tags (EST) were found in the GenBank EST databases by comparing all previously known Arabidopsis miRNAs to GenBank's plant EST databases with BLAST algorithms. After removing the EST sequences with high numbers (more than 2) of mismatched nucleotides, a total of 812 EST contigs were identified. After predicting and scoring the RNA secondary structure of the 812 EST sequences using mFold software, 338 new potential miRNAs were identified in 60 plant species. miRNAs are widespread. Some microRNAs may highly conserve in the plant kingdom, and they may have the same ancestor in very early evolution. There is no nucleotide substitution in most miRNAs among many plant species. Some of the new identified potential miRNAs may be induced and regulated by environmental biotic and abiotic stresses. Some may be preferentially expressed in specific tissues, and are regulated by developmental switching. These findings suggest that EST analysis is a good alternative strategy for identifying new miRNA candidates, their targets, and other genes. A large number of miRNAs exist in different plant species and play important roles in plant developmental switching and plant responses to environmental abiotic and biotic stresses as well as signal transduction. Environmental stresses and developmental switching may be the signals for synthesis and regulation of miRNAs in plants. A model for miRNA induction and expression, and gene regulation by miRNA is hypothesized.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bartel B, Bartel DP . MicroRNAs: at the root of plant development? Plant Physiol 2003; 132:709–17.
Bartel DP . MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97.
Mallory AC, Vaucheret H . MicroRNAs: something important between the genes. Curr Opin Plant Biol 2004; 7:120–5.
Carrington JC, Ambros V . Role of microRNAs in plant and animal development. Science 2003; 301:336–8.
Hunter C, Poethig RS . Missing links: miRNAs and plant development. Curr Opin Genet Dev 2003; 13:372–8.
Griffiths-Jones S . The microRNA Registry. Nucleic Acids Res 2004; 32:D109–11.
Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MCP . microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 2004; 428:84–8.
McHale NA, Koning RE . MicroRNA-directed cleavage of Nicotiana sylvestris PHAVOLUTA mRNA regulates the vascular cambium and structure of apical meristems. Plant Cell 2004; 16:1730–40.
Lai EC . microRNAs: Runts of the genome assert themselves. Curr Biol 2003; 13:R925–36.
Lai EC, Tomancak P, Williams RW, Rubin GM . Computational identification of Drosophila microRNA genes. Genome Biol 2003; 4:R42.
Sunkar R, Zhu JK . Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004; 16:2001–19.
Adai A, Johnson C, Mlotshwa S, et al. Computational prediction of miRNAs in Arabidopsis thaliana. Genome Res 2005; 15:78–91.
Bonnet E, Wuyts J, Rouze P, Van de Peer Y . Detection of 91 potential in plant conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc Natl Acad Sci U S A 2004; 101:11511–6.
Jones-Rhoades MW, Bartel DP . Computational identification of plant MicroRNAs and their targets, including a stress-induced miRNA. Mol Cell 2004; 14:787–99.
Lai EC . Predicting and validating microRNA targets. Genome Biol 2004; 5:A115.
Li Y, Li W, Jin YX . Computational identification of novel family members of microRNA genes in Arabidopsis thaliana and Oryza sativa. Acta Biochim Biophys Sin 2005; 37:75–87.
Rhoades MW, Reinhart BJ, Lim LP, et al. Prediction of plant microRNA targets. Cell 2002; 110:513–20.
Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP . Vertebrate microRNA genes. Science 2003; 299:1540.
Lim LP, Lau NC, Weinstein EG, et al. The microRNAs of Caenorhabditis elegans. Genes Dev 2003; 17:991–1008.
Krol J, Krzyzosiak WJ . Structural aspects of microRNA biogenesis. IUBMB Life 2004; 56:95–100.
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB . Prediction of mammalian microRNA targets. Cell 2003; 115:787–98.
Rajewsky N, Socci ND . Computational identification of microRNA targets. Dev Biol 2004; 267:529–35.
Wang JF, Zhou H, Chen YQ, Luo QJ, Qu LH . Identification of 20 microRNAs from Oryza sativa. Nucleic Acids Res 2004; 32:1688–95.
Wang XJ, Reyes JL, Chua NH, Gaasterland T . Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biology 2004; 5:R65.
Adams MD, Kelley JM, Gocayne JD, et al. Complementary DNA sequencing: expressed sequence tags and human genome project. Science 1991; 252:1651–6.
Matukumalli LK, Grefenstette JJ, Sonstegard TS, van Tassell CP . EST-PAGE - managing and analyzing EST data. Bioinformatics 2004; 20:286–8.
Graham MA, Silverstein KAT, Cannon SB, VandenBosch KA . Computational identification and characterization of novel genes from legumes. Plant Physiol 2004; 135:1179–97.
Jung JD, Park HW, Hahn Y, et al. Discovery of genes for ginsenoside biosynthesis by analysis of ginseng expressed sequence tags. Plant Cell Rep 2003; 22:224–30.
Ohlrogge J, Benning C . Unraveling plant metabolism by EST analysis. Curr Opin Plant Biol 2000; 3:224–8.
Boguski MS, Lowe TM, Tolstoshev CM . dbEST—database for “expressed sequence tags”. Nat Genet 1993; 4:332–3.
Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997; 25:3389–402.
Mathews DH, Sabina J, Zuker M, Turner DH . Expanded sequence dependence of thermodynamic parameters improves prediction of rna secondary structure. J Mol Biol 1999; 288:911–40.
Zuker M . Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406–15.
Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP . MicroRNAs in plants. Genes Dev 2002; 16:1616–26.
Ambros V . MicroRNA pathways in flies and worms: Growth, death, fat, stress, and timing. Cell 2003; 113:673–6.
Smalheiser NR . EST analyses predict the existence of a population of chimeric microRNA precursor-mRNA transcripts expressed in normal human and mouse tissues. Genome Biol 2003; 4:403.
Achard P, Herr A, Baulcombe DC, Harberd NP . Modulation of floral development by a gibberellin-regulated microRNA. Development 2004; 131:3357–65.
Chen XM . A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004; 303:2022–5.
Llave C, Xie ZX, Kasschau KD, Carrington JC . Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002; 297:2053–6.
Palatnik JF, Allen E, Wu XL, et al. Control of leaf morphogenesis by microRNAs. Nature 2003; 425:257–63.
Park W, Li JJ, Song RT, Messing J, Chen XM . CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol 2002; 12:1484–95.
The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 2000; 408:796–815.
Xu PZ, Vernooy SY, Guo M, Hay BA . The Drosophila MicroRNA mir-14 suppresses cell death and is required for normal fat metabolism. Current Biology 2003; 13:790–5.
Llave C . MicroRNAs: more than a role in plant development? Mol Plant Path 2004; 5:361–6.
Aukerman MJ, Sakai H . Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003; 15:2730–41.
Bartel DP, Chen CZ . Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004; 5:396–400.
Zhu JK. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 2002; 53:247–73.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
ZHANG, B., PAN, X., WANG, Q. et al. Identification and characterization of new plant microRNAs using EST analysis. Cell Res 15, 336–360 (2005). https://doi.org/10.1038/sj.cr.7290302
Received:
Revised:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/sj.cr.7290302
Keywords
This article is cited by
-
Machine learning provides specific detection of salt and drought stresses in cucumber based on miRNA characteristics
Plant Methods (2023)
-
Identification of conserved miRNAs and their targets in Jatropha curcas: an in silico approach
Journal of Genetic Engineering and Biotechnology (2023)
-
Characterization of Two-Component System gene (TCS) in melatonin-treated common bean under salt and drought stress
Physiology and Molecular Biology of Plants (2023)
-
Identification and functional analysis of miR156 family and its target genes in foxtail millet (Setaria italica)
Plant Growth Regulation (2023)
-
Comprehensive, integrative genomic analysis of microRNA expression profiles in different tissues of two wheat cultivars with different traits
Functional & Integrative Genomics (2023)