Abstract
Aim:
To study the conformational changes of Aβ42 and discover novel inhibitors of both Aβ42 aggregation and β-secretase (BACE1).
Methods:
A molecular dynamics (MD) simulation at a microsecond level was performed to explore stable conformations of Aβ42 monomer in aqueous solution. Subsequently, structure-based virtual screening was used to search for inhibitors of both Aβ42 aggregation and BACE1. Protein purification and in vitro activity assays were performed to validate the inhibition of the compounds identified via virtual screening.
Results:
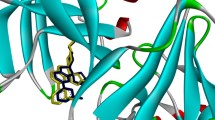
The initial α-helical conformation of Aβ42, which was unstable in aqueous solution, turned into a β-sheet mixed with a coil structure through a transient and fully random coil. The conformation of Aβ42 mainly comprising β-sheets and coils structure was used for further virtual screening. Five compounds were identified as inhibitors for Aβ42 aggregation, and one of them, AE-848, was discovered to be a dual inhibitor of both Aβ42 aggregation and BACE1, with IC50 values of 36.95 μmol/L and 22.70 μmol/L, respectively.
Conclusion:
A helical to β-sheet conformational change in Aβ42 occurred in a 1.8 microsecond MD simulation. The resulting β-sheet structure of the peptide is an appropriate conformation for the virtual screening of inhibitors against Aβ42 aggregation. Five compounds were identified as inhibitors of Aβ42 aggregation by in vitro activity assays. It was particularly interesting to discover a dual inhibitor that targets both Aβ42 aggregation and BACE1, the two crucial players in the pathogenesis of Alzheimer's disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Selkoe DJ . The maolecular pathology of Alzheimer's disease. Neuron 1991; 6: 487–98.
Miller DL, Papayannopoulos IA, Styles J, Bobin SA, Lin YY, Biemann K, et al. Peptide compositions of the cerebrovascular and senile plaque core amyloid deposits of Alzheimer's disease. Arch Biochem Biophys 1993; 301: 41–52.
Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, et al. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature 1987; 325: 733–6.
Mattson MP . Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol Rev 1997; 77: 1081–132.
Selkoe DJ . Alzheimer's disease: genes, proteins, and therapy. Physiol Rev 2001; 81: 741–66.
Hardy J, Selkoe DJ . The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics. Science 2002; 297: 353–6.
Speretta E, Jahn TR, Tartaglia GG, Favrin G, Barros TP, Imarisio S, et al. Expression in drosophila of tandem amyloid beta peptides provides insights into links between aggregation and neurotoxicity. J Biol Chem 2012; 287: 20748–54.
Xia W . Brain amyloid beta protein and memory disruption in Alzheimer's disease. Neuropsychiatr Dis Treat 2010; 6: 605–11.
Liu D, Xu Y, Feng Y, Liu H, Shen X, Chen K, et al. Inhibitor discovery targeting the intermediate structure of beta-amyloid peptide on the conformational transition pathway: implications in the aggregation mechanism of beta-amyloid peptide. Biochemistry 2006; 45: 10963–72.
Vassar R . BACE1: the beta-secretase enzyme in Alzheimer's disease. J Mol Neurosci 2004; 23: 105–14.
Xu Y, Li MJ, Greenblatt H, Chen W, Paz A, Dym O, et al. Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations. Acta Crystallogr D Biol Crystallogr 2012; 68: 13–25.
Rueeger H, Lueoend R, Rogel O, Rondeau JM, Mobitz H, Machauer R, et al. Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure-based design and in vivo reduction of amyloid beta-peptides. J Med Chem 2012; 55: 3364–86.
Mandal M, Zhu Z, Cumming JN, Liu X, Strickland C, Mazzola RD, et al. Design and validation of bicyclic iminopyrimidinones as beta amyloid cleaving enzyme-1 (BACE1) inhibitors: conformational constraint to favor a bioactive conformation. J Med Chem 2012; 55: 9331–45.
Zhou P, Li Y, Fan Y, Wang Z, Chopra R, Olland A, et al. Pyridinyl aminohydantoins as small molecule BACE1 inhibitors. Bioorg Med Chem Lett 2010; 20: 2326–9.
Shimizu H, Tosaki A, Kaneko K, Hisano T, Sakurai T, Nukina N . Crystal structure of an active form of BACE1, an enzyme responsible for amyloid beta protein production. Mol Cell Biol 2008; 28: 3663–71.
Malamas MS, Barnes K, Hui Y, Johnson M, Lovering F, Condon J, et al. Novel pyrrolyl 2-aminopyridines as potent and selective human beta-secretase (BACE1) inhibitors. Bioorg Med Chem Lett 2010; 20: 2068–73.
Coles M, Bicknell W, Watson AA, Fairlie DP, Craik DJ . Solution structure of amyloid beta-peptide (1–40) in a water-micelle environment. Is the membrane-spanning domain where we think it is? Biochemistry 1998; 37: 11064–77.
Xu Y, Shen J, Luo X, Zhu W, Chen K, Ma J, et al. Conformational transition of amyloid beta-peptide. Proc Natl Acad Sci U S A 2005; 102: 5403–7.
Hess B, Kutzner C, van der Spoel D, Lindahl E . GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. journal of chemical theory and computation. J Chem Theory Comput 2008; 4: 435–47.
Oostenbrink C, Villa A, Mark AE, van Gunsteren WF . A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. J Comput Chem 2004; 25: 1656–76.
Kabsch W, Sander C . Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 1983; 22: 2577–637.
Ewing TJ, Makino S, Skillman AG, Kuntz ID . DOCK 4.0: search strategies for automated molecular docking of flexible molecule databases. J Comput Aided Mol Des 2001; 15: 411–28.
Tomaselli S, Esposito V, Vangone P, van Nuland NA, Bonvin AM, Guerrini R, et al. The alpha-to-beta conformational transition of Alzheimer's Abeta-(1–42) peptide in aqueous media is reversible: a step by step conformational analysis suggests the location of beta conformation seeding. Chembiochem 2006; 7: 257–67.
Zhang S, Iwata K, Lachenmann MJ, Peng JW, Li S, Stimson ER, et al. The Alzheimer's peptide a beta adopts a collapsed coil structure in water. J Struct Biol 2000; 130: 130–41.
Stroud JC, Liu C, Teng PK, Eisenberg D . Toxic fibrillar oligomers of amyloid-beta have cross-beta structure. Proc Natl Acad Sci U S A 2012; 109:7717–22.
Ito M, Johansson J, Stromberg R, Nilsson L . Unfolding of the amyloid beta-peptide central helix: mechanistic insights from molecular dynamics simulations. PLoS One 2011; 6: e17587.
Wallace AC, Laskowski RA, Thornton JM . LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 1995; 8: 127–34.
Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A . A partially folded structure of amyloid-beta(1–40) in an aqueous environment. Biochem Biophys Res Commun 2011; 411: 312–6.
Streltsov VA, Varghese JN, Masters CL, Nuttall SD . Crystal structure of the amyloid-beta p3 fragment provides a model for oligomer formation in Alzheimer's disease. J Neurosci 2011; 31: 1419–26.
Crescenzi O, Tomaselli S, Guerrini R, Salvadori S, D'Ursi AM, Temussi PA, et al. Solution structure of the Alzheimer amyloid beta-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur J Biochem 2002; 269: 5642–8.
Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, et al. 3D structure of Alzheimer's amyloid-beta(1–42) fibrils. Proc Natl Acad Sci U S A 2005; 102: 17342–7.
Yu L, Edalji R, Harlan JE, Holzman TF, Lopez AP, Labkovsky B, et al. Structural characterization of a soluble amyloid beta-peptide oligomer. Biochemistry 2009; 48: 1870–7.
Connelly L, Jang H, Arce FT, Ramachandran S, Kagan BL, Nussinov R, et al. Effects of point substitutions on the structure of toxic Alzheimer's beta-amyloid channels: atomic force microscopy and molecular dynamics simulations. Biochemistry 2012; 51: 3031–8.
Reddy G, Straub JE, Thirumalai D . Influence of preformed Asp23-Lys28 salt bridge on the conformational fluctuations of monomers and dimers of Abeta peptides with implications for rates of fibril formation. J Phys Chem B 2009; 113: 1162–72.
Lam AR, Teplow DB, Stanley HE, Urbanc B . Effects of the Arctic (E22→G) mutation on amyloid beta-protein folding: discrete molecular dynamics study. J Am Chem Soc 2008; 130: 17413–22.
Yang M, Teplow DB . Amyloid beta-protein monomer folding: free-energy surfaces reveal alloform-specific differences. J Mol Biol 2008; 384: 450–64.
Sievers SA, Karanicolas J, Chang HW, Zhao A, Jiang L, Zirafi O, et al. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 2011; 475: 96–100.
Kroth H, Ansaloni A, Varisco Y, Jan A, Sreenivasachary N, Rezaei-Ghaleh N, et al. Discovery and structure activity relationship of small molecule inhibitors of toxic beta-amyloid-42 fibril formation. J Biol Chem 2012; 287: 34786–800.
Hard T, Lendel C . Inhibition of amyloid formation. J Mol Biol 2012; 421: 441–65.
Bach JP, Dodel R . Naturally occurring autoantibodies against beta-Amyloid. Adv Exp Med Biol 2012; 750: 91–9.
Wang Y, Huang LQ, Tang XC, Zhang HY . Retrospect and prospect of active principles from Chinese herbs in the treatment of dementia. Acta pharmacol Sin 2010; 31: 649–64.
Deane R, Singh I, Sagare AP, Bell RD, Ross NT, LaRue B, et al. A multimodal RAGE-specific inhibitor reduces amyloid beta-mediated brain disorder in a mouse model of Alzheimer disease. J Clin Invest 2012; 122: 1377–92.
Ghosh AK, Brindisi M, Tang J . Developing beta-secretase inhibitors for treatment of Alzheimer's disease. J Neurochem 2012; 120: 71–83.
Acknowledgements
This study was supported by the “100 Talents Project” of CAS (to Ye-chun XU), the State Key Program of Basic Research of China (Grant No 2009CB918501), and the National Natural Science Foundation of China (No 91013010 and 21172233). Computational resources were supported by the National Supercomputing Center in Tianjin (Tianhe-1), the Shanghai Supercomputer Center and the Computer Network Information Center of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, Yy., Li, L., Chen, Tt. et al. Microsecond molecular dynamics simulation of Aβ42 and identification of a novel dual inhibitor of Aβ42 aggregation and BACE1 activity. Acta Pharmacol Sin 34, 1243–1250 (2013). https://doi.org/10.1038/aps.2013.55
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/aps.2013.55
Keywords
This article is cited by
-
Conformation and dynamics of the C-terminal region in human phosphoglycerate mutase 1
Acta Pharmacologica Sinica (2017)