Abstract
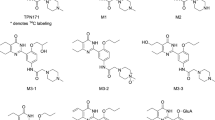
Cryptotanshinone (CT) is the main active component in the root of Salvia miltiorrhiza Bunge (SMB) that displays antibacterial, anti-inflammatory and anticancer activities. In this study, we characterized phase I and phase II metabolism of CT in human liver microsomes in vitro and identified the metabolic enzymes (CYPs and UGTs) involved. The metabolites of CT generated by CYPs were detected using LC-MS/MS and the CYP subtypes involved in the metabolic reactions were identified using chemical inhibitors of CYP enzymes and recombinant human CYP enzymes (CYP1A2, CYP2A6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A4). Glucuronidation of CT was also examined, and the UGT subtypes involved in the metabolic reactions were identified using recombinant human UGT enzymes (1A1, 1A3, 1A4, 1A5, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15 and 2B17). After adding NADPH to the human liver microsomes incubation system, CT was transformed into 6 main dehydrogenation and hydroxylation metabolites. CYP2A6, CYP3A4 and CYP2C19 were the major contributors to the transformation of its hydroxylation metabolites. CYP2C19, CYP1A2 and CYP3A4 were the major contributors to the transformation of its hydrogenation metabolites in human liver microsomes. This study showed that the metabolites at m/z of 473 were mediated by UGT1A9 and that the metabolites at m/z of 489 were mediated by UGT2B7 and UGT2B4. CT was extensively metabolized by UGTs following metabolism by CYPs in the liver.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Cheng TO. Cardiovascular effects of danshen. Int J Cardiol 2007; 121: 9–22.
Lee DS, Lee SH, Noh JG, Hong SD. Antibacterial activities of cryptotanshinone and dihydrotanshinone I from a medicinal herb, Salvia miltiorrhiza Bunge. Biosci Biotechnol Biochem 1999; 63: 2236–9.
Jin DZ, Yin LL, Ji XQ, Zhu XZ. Cryptotanshinone inhibits cyclooxygenase-2 enzyme activity but not its expression. Eur J Pharmacol 2006; 549: 166–72.
Kang BY, Chung SW, Kim SH, Ryu SY, Kim TS. Inhibition of interleukin-12 and interferon-gamma production in immune cells by tanshinones from Salvia miltiorrhiza. Immunopharmacology 2000; 49: 355–61.
Chen W, Liu L, Luo Y, Odaka Y, Awate S, Zhou H, et al. Cryptotanshinone activates p38/JNK and inhibits Erk1/2 leading to caspase-independent cell death in tumor cells. Cancer Prev Res 2012; 5: 778–87.
Xue M, Cui Y, Wang HQ, Luo YJ, Zhang B, Zhou ZT. Pharmacokinetics of cryptotanshinone and its metabolite in pigs. Acta Pharm Sin 1999; 34: 81–4.
Xie MZ, Shen ZF. Absorption, distribution, excretion and metabolism of cryptotanshinone. Acta Pharm Sin 1983; 18: 90–6.
Qiu F, Jiang J, Ma Y, Wang G, Gao C, Zhang X, et al. Opposite effects of single-dose and multidose administration of the ethanol extract of Danshen on CYP3A in healthy volunteers. Evid Based Complement Alternat Med 2013; 730734: 1–8.
Zhang J, Huang M, Guan S, Bi HC, Pan Y, Duan W. A mechanistic study of the intestinal absorption of cryptotanshinone, the major active constituent of Salvia miltiorrhiza. J Pharmacol Exp Ther 2006; 317: 1285–94.
Dai H, Li X, Li X, Bai L, Li Y, Xue M. Coexisted components of Salvia miltiorrhiza enhance intestinal absorption of cryptotanshinone via inhibition of the intestinal P-gp. Phytomedicine 2012; 19: 1256–62.
Gao Y, Shao J, Jiang Z, Chen J, Gu S, Yu S. Drug enterohepatic circulation and disposition: constituents of systems pharmacokinetics. Drug Discov Today 2014; 19: 326–40.
Song M, Hang TJ, Zhang ZX, Du R, Chen J. Determination of cryptotanshinone and its metabolite in rat plasma by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2005; 827: 205–9.
Dai H, Wang M, Li X, Wang L, Li Y, Xue M. Structural elucidation of in vitro and in vivo metabolites of cryptotanshinone by HPLC-DAD-ESI-MS(n). J Pharm Biomed Anal 2008; 48: 885–96.
Liu J, Wu J, Wang X, Cao Z. Study of the phase I and phase II metabolism of a mxture containing multiple tanshinones using liquid chromatography/tandom mass spectrometry. Rapid Commun Mass Spectrom 2007; 21: 2992–8.
Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther 2013; 138: 103–41.
Kiang TK, Ensom MH, Chang TK. UDP-glucuronosyltransferases and clinical drug-drug interactions. Pharmacol Ther 2005; 106: 97–132.
Emoto C, Murayama N, Rostami-Hodjegan A, Yamazaki H. Methodologies for investigating drug metabolism at the early drug discovery stage: prediction of hepatic drug clearance and P450 contribution. Curr Drug Metab 2010; 11: 678–85.
Fan G, Jiang X, Wu X, Fordjour PA, Miao L, Zhang H, et al. Anti-infiammatory activity of tanshinone IIA in LPS-stimulated RAW264.7 macrophages via miRNAs and TLR4-NF-κB pathway. Infiammation 2016; 39: 375–84.
Xie J, Liu J, Liu H, Liang S, Lin M, Gu Y, et al. The antitumor effect of tanshinone IIA on anti-proliferation and decreasing VEGF/VEGFR2 expression on the human non-small cell lung cancer A549 cell line. Acta Pharm Sin B 2015; 5: 554–63.
Joo J, Kim YW, Wu Z, Shin JH, Lee B, Shon JC, et al. Screening of non-steroidal anti-inflammatory drugs for inhibitory effects on the activities of six UDP-glucuronosyltransferases (UGT1A1, 1A3, 1A4, 1A6, 1A9 and 2B7) using LC-MS/MS. Biopharm Drug Dispos 2015; 36: 258–64.
Foti RS, Dalvie DK. Cytochrome P450 and non-cytochrome P450 oxidative metabolism: contributions to the pharmacokinetics, safety, and efficacy of xenobiotics. Drug Metab Dispos 2016; 44: 1229–45.
Wang Q, Hao H, Zhu X, Yu G, Lai L, Liu Y, et al. Regioselect glucuronidation of tanshinone IIA after Quinone reduction: identification of human UDP-glucuronosyltromsferases, species difference and interaction potential. Drug Metab Dispos 2010; 38: 1132–40.
Wu X, Zhang Q, Guo J, Jia Y, Zhang Z, Zhao M, et al. Metabolism of F18, a derivative of calanolide A, in human liver microsomes and cytosol. Front Pharmacol 2017; 8:1–18.
Yang X, Atkinson K, Di L. Novel cytochrome P450 reaction phenotyping for low clearance compounds using the hepatocyte relay method. Drug Metab Dispos 2016; 44: 460–5.
Sun J, Yang M, Han J, Wang B, Ma X, Xu M, et al. Profiling the metabolic difference of seven tanshinones using high-performance liquid chromatography/multi-stage mass spectrometry with data-dependent acquisition. Rapid Commun Mass Spectrom 2007; 21: 2211–26.
Song YL, Jing WH, Yan R, Wang YT. Metabolic characterization of (±)-praeruptorin A in vitro and in vivo by high performance liquid chromatography coupled with hybrid triple quadrupole-linear ion trap mass spectrometry and time-of-flight mass spectrometry. J Pharm Biomed Anal 2014; 90: 98–110.
Liu HX, Hu Y, Liu Y, He YQ, Li W, Yang L. Hydroxylation of tanshinone IIa in human liver microsomes is specifically catalysed by cytochrome P4502A6. Xenobiotica 2009; 39: 382–90.
He P, Court MH, Greenblatt DJ, Von Moltke LL. Genotype-phenotype associations of cytochrome P450 3A4 and 3A5 polymorphism with midazolam clearance in vivo. Clin Pharmacol Ther 2005; 77: 373–87.
Özhan G, Mutur M, Ercan G, Alpertunga B. Genetic variations in the xenobiotic-metabolizing enzymes CYP1A1, CYP1A2, CYP2C9, CYP2C19 and susceptibility to colorectal cancer among Turkish people. Genet Test Mol Biomarkers 2014; 18: 223–8.
Nurfadhlina M, Foong K, The LK, Tan SC, Mohd Zaki S, Ismail R. CYP2A6 polymorphisms in Malays, Chinese and Indians. Xenobiotica 2006; 36: 684–92.
Hu YR, Qiao HL, Kan QC. Pharmacokinetics of lansoprazole in Chinese healthy subjects in relation to CYP2C19 genotypes. Acta Pharmacol Sin 2004; 25: 986–90.
Court MH, Duan SX, von Moltke LL, Greenblatt DJ, Patten CJ, Miners JO. Interindividual variability in acetaminophen glucuronidation by human liver microsomes: identification of relevant acetaminophen UDP-glucuronosyltransferase isoforms. J Pharmacol Exp Ther 2001; 299: 998–1006.
Hwang MS, Lee SJ, Jeong HE, Lee S, Yoo MA, Shin JG. Genetic variations in UDP-glucuronosyltransferase 2B7 gene (UGT2B7) in a Korean population. Drug Metab Pharmacokinet 2010; 25: 398–402.
Yu C, Ye S, Sun H, Liu Y, Gao L, Shen C, et al. PXR-mediated transcriptional activation of CYP3A4 by cryptotanshinone and tanshinone IIA. Chem Biol Interact 2009; 177: 58–64.
Zhang R, Sun J, Ma L, Wu X, Pan G, Hao H. Induction of cytochromes P450 1A1 and 1A2 by tanshinones in human HepG2 hepatoma cell line. Toxicol Appl Pharmacol 2011; 252: 18–27.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No 81173118) and Shanghai Key Lab of Traditional Clinical Medicine (Grant C14dZ2273200).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Zeng, J., Fan, Yj., Tan, B. et al. Charactering the metabolism of cryptotanshinone by human P450 enzymes and uridine diphosphate glucuronosyltransferases in vitro . Acta Pharmacol Sin 39, 1393–1404 (2018). https://doi.org/10.1038/aps.2017.144
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/aps.2017.144
Keywords
This article is cited by
-
Pharmacological impacts of tanshinone on osteogenesis and osteoclastogenesis: a review
Naunyn-Schmiedeberg's Archives of Pharmacology (2025)
-
High fat diet significantly changed the global gene expression profile involved in hepatic drug metabolism and pharmacokinetic system in mice
Nutrition & Metabolism (2020)