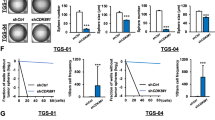
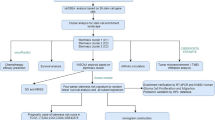
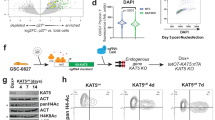
Abstract
Cullin3 E3 ubiquitin ligase ubiquitinates a wide range of substrates through substrate-specific adaptors Bric-a-brac, Tramtrack, and Broad complex (BTB) domain proteins. These E3 ubiquitin ligase complexes are involved in diverse cellular functions. Our recent study demonstrated that decreased Cullin3 expression induces glioma initiation and correlates with poor prognosis of patients with malignant glioma. However, the substrate recognition mechanism associated with tumorigenesis is not completely understood. Through yeast two-hybrid screening, we identified potassium channel tetramerization domain-containing 2 (KCTD2) as a BTB domain protein that binds to Cullin3. The interaction of Cullin3 and KCTD2 was verified using immunoprecipitation and immunofluorescence. Of interest, KCTD2 expression was markedly decreased in patient-derived glioma stem cells (GSCs) compared with non-stem glioma cells. Depletion of KCTD2 using a KCTD2-specific short-hairpin RNA in U87MG glioma cells and primary Ink4a/Arf-deficient murine astrocytes markedly increased self-renewal activity in addition with an increased expression of stem cell markers, and mouse in vivo intracranial tumor growth. As an underlying mechanism for these KCTD2-mediated phenotypic changes, we demonstrated that KCTD2 interacts with c-Myc, which is a key stem cell factor, and causes c-Myc protein degradation by ubiquitination. As a result, KCTD2 depletion acquires GSC features and affects aerobic glycolysis via expression changes in glycolysis-associated genes through c-Myc protein regulation. Of clinical significance was our finding that patients having a profile of KCTD2 mRNA-low and c-Myc gene signature-high, but not KCTD2 mRNA-low and c-Myc mRNA-high, are strongly associated with poor prognosis. This study describes a novel regulatory mode of c-Myc protein in malignant gliomas and provides a potential framework for glioma therapy by targeting c-Myc function.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Hershko A, Ciechanover A . The ubiquitin system. Annu Rev Biochem 1998; 67: 425–479.
Sarikas A, Hartmann T, Pan ZQ . The cullin protein family. Genome Biol 2011; 12: 220.
Genschik P, Sumara I, Lechner E . The emerging family of CULLIN3-RING ubiquitin ligases (CRL3s): cellular functions and disease implications. EMBO J 2013; 32: 2307–2320.
Furukawa M, He YJ, Borchers C, Xiong Y . Targeting of protein ubiquitination by BTB-Cullin 3-Roc1 ubiquitin ligases. Nat Cell Biol 2003; 5: 1001–1007.
Smaldone G, Pirone L, Balasco N, Di Gaetano S, Pedone EM, Vitagliano L . Cullin 3 recognition is not a universal property among KCTD proteins. PLoS One 2015; 10: e0126808.
Wen PY, Kesari S . Malignant gliomas in adults. N Engl J Med 2008; 359: 492–507.
Vescovi AL, Galli R, Reynolds BA . Brain tumor stem cells. Nat Rev Cancer 2006; 6: 425–436.
Stopschinski BE, Beier CP, Beier D . Glioblastoma cancer stem cells – from concept to clinical application. Cancer Lett 2013; 338: 32–40.
Jin X, Jeon HM, Jin X, Kim EJ, Yin J, Jeon HY et al. ID1-Cullin3 axis regulates intracellular SHH and WNT signaling in glioblastoma stem cells. Cell Rep 2016; 16: 1629–1641.
Eilers M, Eisenman RN . Myc’s broad reach. Genes Dev 2008; 22: 2755–2766.
Nesbit CE, Tersak JM, Prochownik EV . MYC oncogenes and human neoplastic disease. Oncogene 1999; 18: 3004–3016.
Wang J, Wang H, Li Z, Wu Q, Lathia JD, McLendon RE et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One 2008; 3: e3769.
Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA 2013; 110: 8644–8649.
Bennett EJ, Rush J, Gygi SP, Harper JW . Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 2010; 143: 951–965.
Singer JD, Gurian-West M, Clurman B, Roberts JM . Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev 1999; 13: 2375–2387.
He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT et al. Identification of c-Myc as a target of the APC pathway. Science 1998; 281: 1509–1512.
Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A et al. Hedgehog signaling in prostate regeneration, neoplasia and metastasis. Nature 2004; 431: 707–712.
Dang CV, Le A, Gao P . MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res 2009; 15: 6479–6483.
Vander Heiden MG, Cantley LC, Thompson CB . Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009; 324: 1029–1033.
Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K . Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res 2009; 7: 157–167.
Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010; 17: 98–110.
Guan X, Vengoechea J, Zheng S, Sloan AE, Chen Y, Brat DJ et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One 2014; 9: e91216.
Thu KL, Pikor LA, Chari R, Wilson IM, Macaulay CE, English JC et al. Genetic disruption of KEAP1/Cul3 E3 ubiquitin ligase complex components is a key mechanism of NF-kappaB pathway activation in lung cancer. J Thorac Oncol 2011; 6: 1521–1529.
Kossatz U, Breuhahn K, Wolf B, Hardtke-Wolenski M, Wilkens L, Steinemann D et al. The cyclin E regulator cullin 3 prevents mouse hepatic progenitor cells from becoming tumor-initiating cells. J Clin Invest 2010; 120: 3820–3833.
Lange S, Perera S, Teh P, Chen J . Obscurin and KCTD6 regulate cullin-dependent small ankyrin-1 (sAnk1.5) protein turnover. Mol Biol Cell 2012; 23: 2490–2504.
De Smaele E, Di Marcotullio L, Moretti M, Pelloni M, Occhione MA, Infante P et al. Identification and characterization of KCASH2 and KCASH3, 2 novel Cullin3 adaptors suppressing histone deacetylase and Hedgehog activity in medulloblastoma. Neoplasia 2011; 13: 374–3850.
Chen Y, Yang Z, Meng M, Zhao Y, Dong N, Yan H et al. Cullin mediates degradation of RhoA through evolutionarily conserved BTB adaptors to control actin cytoskeleton structure and cell movement. Mol Cell 2009; 35: 841–855.
Sobieszczuk DF, Poliakov A, Xu Q, Wilkinson DG . A feedback loop mediated by degradation of an inhibitor is required to initiate neuronal differentiation. Genes Dev 2010; 24: 206–218.
Araki R, Hoki Y, Uda M, Nakamura M, Jincho Y, Tamura C et al. Crucial role of c-Myc in the generation of induced pluripotent stem cells. Stem Cells 2011; 29: 1362–1370.
Faria MH, Khayat AS, Burbano RR, Rabenhorst SH . c -MYC amplification and expression in astrocytic tumors. Acta Neuropathol 2008; 116: 87–95.
Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G et al. A signaling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 2004; 6: 308–318.
Welcker M, Clurman BE . FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 2008; 8: 83–93.
Kim SH, Park J, Choi MC, Kim HP, Park JH, Jung Y et al. Zinc-fingers and homeoboxes 1 (ZHX1) binds DNA methyltransferase (DNMT) 3B to enhance DNMT3B-mediated transcriptional repression. Biochem Biophys Res Commun 2007; 2: 318–323.
Kim SH, Kim EJ, Hitomi M, Oh SY, Jin X, Jeon HM et al. The LIM-only transcription factor LMO2 determines tumorigenic and angiogenic traits in glioma stem cells. Cell death Differ 2015; 22: 1517–1525.
Acknowledgements
We thank all members of Cell Growth Regulation Lab for their helpful discussions and technical assistances. This work was supported by grants from the National Research Foundation (NRF) funded by the Ministry of Science, ICT and Future Planning (2011-0017544, 2015R1A5A1009024 and 2016R1D1A1B03931941), from Next-Generation Biogreen21 Program (PJ01107701), from the General Program of the National Natural Science Foundation of China (no. 81572891), and from Korea University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by JP Medema
Rights and permissions
About this article
Cite this article
Kim, EJ., Kim, SH., Jin, X. et al. KCTD2, an adaptor of Cullin3 E3 ubiquitin ligase, suppresses gliomagenesis by destabilizing c-Myc. Cell Death Differ 24, 649–659 (2017). https://doi.org/10.1038/cdd.2016.151
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cdd.2016.151
This article is cited by
-
Endothelial cells-derived SEMA3G suppresses glioblastoma stem cells by inducing c-Myc degradation
Cell Death & Differentiation (2025)
-
Proteostasis and Its Role in Disease Development
Cell Biochemistry and Biophysics (2024)
-
E3 ubiquitin ligases in cancer stem cells: key regulators of cancer hallmarks and novel therapeutic opportunities
Cellular Oncology (2023)
-
The emerging role of the KCTD proteins in cancer
Cell Communication and Signaling (2021)
-
Cul3 is required for normal development of the mammary gland
Cell and Tissue Research (2021)