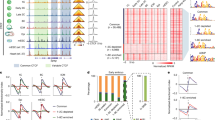
Abstract
Oocytes display a maternal-specific gene expression profile, which is switched to a zygotic profile when a haploid set of chromatin is passed on to the fertilized egg that develops into an embryo. The mechanism underlying this transcription reprogramming is currently unknown. Here we demonstrate that by the time when transcription is shut down in germinal vesicle oocytes, a range of general transcription factors and transcriptional regulators are dissociated from the chromatin. The global dissociation of chromatin factors (CFs) disrupts physical contacts between the chromatin and CFs and leads to erasure of the maternal transcription program at the functional level. Critical transcription factors and regulators remain separated from chromatin for a prolonged period, and become re-associated with chromatin shortly after pronuclear formation. This is followed temporally by the re-establishment of nuclear functions such as DNA replication and transcription. We propose that the maternal transcription program is erased during oogenesis to generate a relatively naïve chromatin and the zygotic transcription program is rebuilt de novo after fertilization. This process is termed as the “erase-and-rebuild” process, which is used to reset the transcription program, and most likely other nuclear processes as well, from a maternal one to that of the embryo. We further show in the accompanying paper (Gao T, et al., Cell Res 2007; 17: 135-150.) that the same strategy is also employed to reprogram transcriptional profiles in somatic cell nuclear transfer and parthenogenesis, suggesting that this model is universally applicable to all forms of transcriptional reprogramming during early embryogenesis. Displacement of CFs from chromatin also offers an explanation for the phenomenon of transcription silence during the maternal to zygotic transition.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Bouniol-Baly C, Hamraoui L, Guibert J, Beaujean N, Szollosi MS, Debey P . Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol Reprod 1999; 60:580–587.
De La Fuente R, Viveiros MM, Burns KH, Adashi EY, Matzuk MM, Eppig JJ . Major chromatin remodeling in the germinal vesicle (GV) of mammalian oocytes is dispensable for global transcriptional silencing but required for centromeric heterochromatin function. Dev Biol 2004; 275:447–458.
De La Fuente R . Chromatin modifications in the germinal vesicle (GV) of mammalian oocytes. Dev Biol 2006; 292:1–12.
De La Fuente R, Eppig JJ . Transcriptional activity of the mouse oocyte genome: companion granulosa cells modulate transcription and chromatin remodeling. Dev Biol 2001; 229:224–236.
Miyara F, Migne C, Dumont-Hassan M, et al. Chromatin configuration and transcriptional control in human and mouse oocytes. Mol Reprod Dev 2003; 64:458–470.
Ram PT, Schultz RM . Reporter gene expression in G2 of the 1-cell mouse embryo. Dev Biol 1993; 156:552–556.
Aoki F, Worrad DM, Schultz RM . Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181:296–307.
Schultz RM . The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update 2002; 8:323–331.
Bouniol C, Nguyen E, Debey P . Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res 1995; 218:57–62.
Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T . Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev 2006; 20:1744–1754.
Surani MA . Nuclear reprogramming by human embryonic stem cells. Cell 2005; 122:653–654.
Hochedlinger K, Jaenisch R . Nuclear reprogramming and pluripotency. Nature 2006; 441:1061–1067.
Gurdon JB, Byrne JA . The first half-century of nuclear transplantation. Biosci Rep 2004; 24:545–557.
Edgar BA, Schubiger G . Parameters controlling transcriptional activation during early Drosophila development. Cell 1986; 44:871–877.
Kimelman D, Kirschner M, Scherson T . The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell 1987; 48:399–407.
Yasuda GK, Schubiger G . Temporal regulation in the early embryo: is MBT too good to be true? Trends Genet 1992; 8:124–127.
Newport J, Kirschner M . A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell 1982; 30:687–696.
Prioleau MN, Huet J, Sentenac A, Mechali M . Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 1994; 77:439–449.
Prioleau MN, Buckle RS, Mechali M . Programming of a repressed but committed chromatin structure during early development. EMBO J 1995; 14:5073–5084.
Almouzni G, Wolffe AP . Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. EMBO J 1995; 14:1752–1765.
Bell P, Scheer U . Developmental changes in RNA polymerase I and TATA box-binding protein during early Xenopus embryogenesis. Exp Cell Res 1999; 248:122–135.
Veenstra GJ, Destree OH, Wolffe AP . Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Mol Cell Biol 1999; 19:7972–7982.
Worrad DM, Ram PT, Schultz RM . Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development 1994; 120:2347–2357.
Parfenov VN, Pochukalina GN, Davis DS, Reinbold R, Scholer HR, Murti KG . Nuclear distribution of Oct-4 transcription factor in transcriptionally active and inactive mouse oocytes and its relation to RNA polymerase II and splicing factors. J Cell Biochem 2003; 89:720–732.
Parfenov VN, Davis DS, Pochukalina GN, Kostyuchek D, Murti KG . Nuclear distribution of RNA polymerase II in human oocytes from antral follicles: dynamics relative to the transcriptional state and association with splicing factors. J Cell Biochem 2000; 77:654–665.
Bellier S, Chastant S, Adenot P, Vincent M, Renard JP, Bensaude O . Nuclear translocation and carboxyl-terminal domain phosphorylation of RNA polymerase II delineate the two phases of zygotic gene activation in mammalian embryos. EMBO J 1997; 16:6250–6262.
Gebara MM, Sayre MH, Corden JL . Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J Cell Biochem 1997; 64:390–402.
Miyagishima H, Isono K, Fujimura Y, et al. Dissociation of mammalian Polycomb-group proteins, Ring1B and Rae28/Ph1, from the chromatin correlates with configuration changes of the chromatin in mitotic and meiotic prophase. Histochem Cell Biol 2003; 120:111–119.
Zatsepina OV, Bouniol-Baly C, Amirand C, Debey P . Functional and molecular reorganization of the nucleolar apparatus in maturing mouse oocytes. Dev Biol 2000; 223:354–370.
Kornberg RD . Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 2005; 30:235–239.
Lemon B, Tjian R . Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 2000; 14:2551–2569.
Orphanides G, Reinberg D . A unified theory of gene expression. Cell 2002; 108:439–451.
Green MR . Eukaryotic transcription activation: right on target. Mol Cell 2005; 18:399–402.
Bouniol-Baly C, Nguyen E, Besombes D, Debey P . Dynamic organization of DNA replication in one-cell mouse embryos: relationship to transcriptional activation. Exp Cell Res 1997; 236:201–211.
Adenot PG, Mercier Y, Renard JP, Thompson EM . Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development 1997; 124:4615–4625.
Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I . An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86:679–688.
Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R . Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 1998; 394:369–374.
Debey P, Szollosi MS, Szollosi D, Vautier D, Girousse A, Besombes D . Competent mouse oocytes isolated from antral follicles exhibit different chromatin organization and follow different maturation dynamics. Mol Reprod Dev 1993; 36:59–74.
Mattson BA, Albertini DF . Oogenesis: chromatin and microtubule dynamics during meiotic prophase. Mol Reprod Dev 1990; 25:374–83.
Wickramasinghe D, Ebert KM, Albertini DF . Meiotic competence acquisition is associated with the appearance of M-phase characteristics in growing mouse oocytes. Dev Biol 1991; 143:162–172.
Zuccotti M, Piccinelli A, Giorgi Rossi P, Garagna S, Redi CA . Chromatin organization during mouse oocyte growth. Mol Reprod Dev 1995; 41:479–485.
Kim JM, Ogura A, Nagata M, Aoki F . Analysis of the mechanism for chromatin remodeling in embryos reconstructed by somatic nuclear transfer. Biol Reprod 2002; 67:760–766.
Yang Y, Cao J, Huang L, Fang HY, Sheng HZ . Regulated expression of TATA-binding protein-related factor 3 (TRF3) during early embryogenesis. Cell Res 2006; 16:610–621.
Kadonaga JT . Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 2004; 116:247–257.
Soutoglou E, Talianidis I . Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 2002; 295:1901–1904.
Fry CJ, Peterson CL . Transcription. Unlocking the gates to gene expression. Science 2002; 295:1847–1848.
Thomas MJ, Seto E . Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene 1999; 236:197–208.
Varga-Weisz PD, Becker PB . Chromatin-remodeling factors: machines that regulate? Curr Opin Cell Biol 1998; 10:346–353.
Tate P, Skarnes W, Bird A . The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet 1996; 12:205–208.
Bakshi RP, Galande S, Muniyappa K . Functional and regulatory characteristics of eukaryotic type II DNA topoisomerase. Crit Rev Biochem Mol Biol 2001; 36:1–37.
Maison C, Almouzni G . HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 2004; 5:296–304.
Mondal N, Parvin JD . DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature 2001; 413:435–438.
Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M . Mitotic remodeling of the replicon and chromosome structure. Cell 2005; 123:787–801.
Chen D, Hinkley CS, Henry RW, Huang S . TBP dynamics in living human cells: constitutive association of TBP with mitotic chromosomes. Mol Biol Cell 2002; 13:276–284.
Chen D, Dundr M, Wang C, et al. Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J Cell Biol 2005; 168:41–54.
Wang K, Sun F, Sheng HZ . Regulated expression of TAF1 in 1-cell mouse embryos. Zygote 2006; 14:209–215.
Sun F, Tang F, Yan AY, Fang HY, Sheng HZ . Expression of SRG3, a chromatin remodeling factor, in the mouse oocyte and early preimplantation embryos. Zygote 2007; 15:1–10.
Luthardt FW, Donahue RP . Pronuclear DNA synthesis in mouse eggs. An autoradiographic study. Exp Cell Res 1973; 82:143–151.
Schatten G, Simerly C, Palmer DK, et al. Kinetochore appearance during meiosis, fertilization and mitosis in mouse oocytes and zygotes. Chromosoma 1988; 96:341–352.
Davidson EH . Gene Activity in Early Development. Orlando, FL: Academic Press, 1986.
Kelly WG, Schaner CE, Dernburg AF, et al. X-chromosome silencing in the germline of C. elegans. Development 2002; 129:479–492.
Schisa JA, Pitt JN, Priess JR . Analysis of RNA associated with P granules in germ cells of C. elegans adults. Development 2001; 128:1287–1298.
Taylor JH . Nucleic acid synthesis in relation to the cell division cycle. Ann NY Acad Sci 1960; 90:409–421.
Prescott DM, Bender MA . Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res 1962; 26:260–268.
Littau VC, Allfrey VG, Frenster JH, Mirsky AE . Active and inactive regions of nuclear chromatin as revealed by electron microscope autoradiography. Proc Natl Acad Sci USA 1964; 52:93–100.
Johnson TC, Holland JJ . Ribonucleic acid and protein synthesis in mitotic HeLa cells. J Cell Biol 1965; 27:565–574.
Reik W, Dean W, Walter J . Epigenetic reprogramming in mammalian development. Science 2001; 293:1089–1093.
Santos F, Zakhartchenko V, Stojkovic M, et al. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol 2003; 13:1116–1121.
Surani MA . Reprogramming of genome function through epigenetic inheritance. Nature 2001; 414:122–128.
Kim JM, Liu H, Tazaki M, Nagata M, Aoki F . Changes in histone acetylation during mouse oocyte meiosis. J Cell Biol 2003; 162:37–46.
Gao T, Zheng J, Xing F, et al. Nuclear reprogramming: the strategy used in normal development is also used in somatic cell nuclear transfer and parthenogenesis. Cell Res 2007; 17: 135–150.
Imhof A, Wolffe AP . Transcription: gene control by targeted histone acetylation. Curr Biol 1998; 8:R422–R424.
Martianov I, Viville S, Davidson I . RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science 2002; 298:1036–1039.
Acknowledgements
We are grateful to Drs Yun-Bo Shi, Yingzi Yang, and Paul Zhou for criticizing the manuscript, Dr Shangang Li for assistance in nuclear transfer procedure, Wei Su for art work, Youming Zhu for cell culture, and Xun Gong, Hui Ding, and Wei Liu for assistance in immunochemistry, Fengying Li and Wanli Li for animal care, and Ayong Yan for nuclear transfer into MII oocytes. We also apologize for citing reviews instead of original publications in some places due to space limitations. Please look in the reviews for original papers.
The study was supported by grants from National Basic Research Program of China (973 Program) (No. 001CB509903, 001CB509904), Hi-Tech Research and Development Program of China (863 Program) (No. 2001AA216121, 2004AA205010), Science and Technology Committee of Shanghai Municipality (No. 99DJ14002, 00DJ1 4033, 01DJ14003, 03DJ14017), Shanghai Municipal Education Commission (No. T0205) and Shanghai Jiao Tong University, School of Medicine.
Contributions: Ruizhen Li is responsible for most analyses in GV oocytes, Feng Sun and Junke Zheng for analyses in one-cell embryos, Tianlong Gao and Haiyan Fang for GFP-CF analyses, Xuejin Chen and Wenqin Ying for nuclear transfer procedures. Feng Sun assisted in manuscript writing. Hui Z Sheng is responsible for development of the model, project planning, and most manuscript writing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, F., Fang, H., Li, R. et al. Nuclear reprogramming: the zygotic transcription program is established through an “erase-and-rebuild” strategy. Cell Res 17, 117–134 (2007). https://doi.org/10.1038/cr.2007.1
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2007.1
Keywords
This article is cited by
-
Epigenetic modification with trichostatin A does not correct specific errors of somatic cell nuclear transfer at the transcriptomic level; highlighting the non-random nature of oocyte-mediated reprogramming errors
BMC Genomics (2016)
-
Oocyte-expressed yes-associated protein is a key activator of the early zygotic genome in mouse
Cell Research (2016)
-
The inability of fully grown germinal vesicle stage oocyte cytoplasm to transcriptionally silence transferred transcribing nuclei
Histochemistry and Cell Biology (2009)
-
Mediators of reprogramming: transcription factors and transitions through mitosis
Nature Reviews Molecular Cell Biology (2008)
-
PRC1 and Suv39h specify parental asymmetry at constitutive heterochromatin in early mouse embryos
Nature Genetics (2008)