Abstract
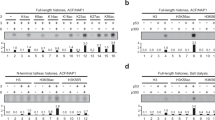
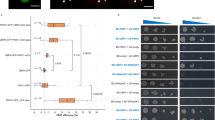
Chromatin structure is important for controlling gene expression, but mechanisms underlying chromatin remodeling are not fully understood. Here we report that an FKBP (FK506 binding protein) type immunophilin, AtFKBP53, possesses histone chaperone activity and is required for repressing ribosomal gene expression in Arabidopsis. The AtFKBP53 protein is a multidomain FKBP with a typical peptidylprolyl isomerase (PPIase) domain and several highly charged domains. Using nucleosome assembly assays, we showed that AtFKBP53 has histone chaperone activity and the charged acidic domains are sufficient for the activity. We show that AtFKBP53 interacts with histone H3 through the acidic domains, whereas the PPIase domain is dispensable for histone chaperone activity or histone binding. Ribosomal RNA gene (18S rDNA) is overexpressed when AtFKBP53 activity is reduced or eliminated in Arabidopsis plants. Chromatin immunoprecipitation assay showed that AtFKBP53 is associated with the 18S rDNA gene chromatin, implicating that AtFKBP53 represses rRNA genes at the chromatin level. This study identifies a new histone chaperone in plants that functions in chromatin remodeling and regulation of transcription.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bentley GA, Finch JT, Lewit-Bentley A, Roth M . The crystal structure of the nucleosome core particle by contrast variation. Basic Life Sci 1984; 27:105–117.
Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A . Structure of the nucleosome core particle at 7 A resolution. Nature 1984; 311:532–537.
Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ . Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997; 389:251–260.
Dutta S, Akey IV, Dingwall C, et al. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol Cell 2001; 8:841–853.
Ito T, Bulger M, Kobayashi R, Kadonaga JT . Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol Cell Biol 1996; 16:3112–3124.
Rodriguez P, Munroe D, Prawitt D, et al. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics 1997; 44:253–265.
Kuzuhara T, Horikoshi M . A nuclear FK506-binding protein is a histone chaperone regulating rDNA silencing. Nat Struct Mol Biol 2004; 11:275–283.
Loppin B, Bonnefoy E, Anselme C, et al. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 2005; 437:1386–1390.
Adams CR, Kamakaka RT . Chromatin assembly: biochemical identities and genetic redundancy. Curr Opin Genet Dev 1999; 9:185–190.
Angelov D, Bondarenko VA, Almagro S, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. Embo J 2006; 25:1669–1679.
Zhu Y, Dong A, Meyer D, et al. Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 2006; 18:2879–2892.
Taneva SG, Banuelos S, Falces J, et al. A mechanism for histone chaperoning activity of nucleoplasmin: thermodynamic and structural models. J Mol Biol 2009; 393:448–463.
Daganzo SM, Erzberger JP, Lam WM, et al. Structure and function of the conserved core of histone deposition protein Asf1. Curr Biol 2003; 13:2148–2158.
Linger J, Tyler JK . The yeast histone chaperone chromatin assembly factor 1 protects against double-strand DNA-damaging agents. Genetics 2005; 171:1513–1522.
Kaya H, Shibahara KI, Taoka KI, et al. FASCIATA genes for chromatin assembly factor-1 in Arabidopsis maintain the cellular organization of apical meristems. Cell 2001; 104:131–142.
March-Diaz R, Reyes JC . The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2009; 2:565–577.
Choi K, Park C, Lee J, et al. Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 2007; 134:1931–1941.
Natsume R, Eitoku M, Akai Y, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature 2007; 446:338–341.
Park YJ, Luger K . The structure of nucleosome assembly protein 1. Proc Natl Acad Sci USA 2006; 103:1248–1253.
Liu ZQ, Gao J, Dong AW, Shen WH . A truncated Arabidopsis NUCLEOSOME ASSEMBLY PROTEIN 1, AtNAP1;3T, alters plant growth responses to abscisic acid and salt in the Atnap1;3–2 mutant. Mol Plant 2009; 2:688–699.
Alexandre C, Moller-Steinbach Y, Schonrock N, Gruissem W, Hennig L . Arabidopsis MSI1 is required for negative regulation of the response to drought stress. Mol Plant 2009; 2:675–687.
Shen WH, Xu L . Chromatin remodeling in stem cell maintenance in Arabidopsis thaliana. Mol Plant 2009; 2:600–609.
He Y . Control of the transition to flowering by chromatin modifications. Mol Plant 2009; 2:554–564.
Georgel PT . Chromatin structure of eukaryotic promoters: a changing perspective. Biochem Cell Biol 2002; 80:295–300.
Eitoku M, Sato L, Senda T, Horikoshi M . Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci 2008; 65:414–444.
Adkins MW, Howar SR, Tyler JK . Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell 2004; 14:657–666.
Korber P, Barbaric S, Luckenbach T, et al. The histone chaperone Asf1 increases the rate of histone eviction at the yeast PHO5 and PHO8 promoters. J Biol Chem 2006; 281:5539–5545.
Zabaronick SR, Tyler JK . The histone chaperone anti-silencing function 1 is a global regulator of transcription independent of passage through S phase. Mol Cell Biol 2005; 25:652–660.
Shan X, Xue Z, Melese T . Yeast NPI46 encodes a novel prolyl cis-trans isomerase that is located in the nucleolus. J Cell Biol 1994; 126:853–862.
Xiao H, Jackson V, Lei M . The FK506-binding protein, Fpr4, is an acidic histone chaperone. FEBS Lett 2006; 580:4357–4364.
Li H, He Z, Lu G, et al. A WD40 domain cyclophilin interacts with histone H3 and functions in gene repression and organogenesis in Arabidopsis. Plant Cell 2007; 19:2403–2416.
He Z, Li L, Luan S . Immunophilins and parvulins. Superfamily of peptidyl prolyl isomerases in Arabidopsis. Plant Physiol 2004; 134:1248–1267.
Okuwaki M, Matsumoto K, Tsujimoto M, Nagata K . Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett 2001; 506:272–276.
Erard MS, Belenguer P, Caizergues-Ferrer M, Pantaloni A, Amalric F . A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur J Biochem 1988; 175:525–530.
Shintomi K, Iwabuchi M, Saeki H, et al. Nucleosome assembly protein-1 is a linker histone chaperone in Xenopus eggs. Proc Natl Acad Sci USA 2005; 102:8210–8215.
Lee IS, Oh SM, Kim SM, Lee DS, Seo SB . Highly acidic C-terminal domain of pp32 is required for the interaction with histone chaperone, TAF-Ibeta. Biol Pharm Bull 2006; 29:2395–2398.
Silverman BD . Hydrophobic and acidic moments of a nucleoplasmin NP-core chaperone. J Biomol Struct Dyn 2006; 24:49–56.
Oeda K, Salinas J, Chua NH . A tobacco bZip transcription activator (TAF-1) binds to a G-box-like motif conserved in plant genes. EMBO J 1991; 10:1793–1802.
Ishimi Y, Kikuchi A . Identification and molecular cloning of yeast homolog of nucleosome assembly protein I which facilitates nucleosome assembly in vitro. J Biol Chem 1991; 266:7025–7029.
Smith S, Stillman B . Stepwise assembly of chromatin during DNA replication in vitro. EMBO J 1991; 10:971–980.
Terakura S, Ueno Y, Tagami H, et al. An oncoprotein from the plant pathogen agrobacterium has histone chaperone-like activity. Plant Cell 2007; 19:2855–2865.
Ori N, Eshed Y, Chuck G, Bowman JL, Hake S . Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 2000; 127:5523–5532.
Gendrel AV, Lippman Z, Yordan C, Colot V, Martienssen RA . Dependence of heterochromatic histone H3 methylation patterns on the Arabidopsis gene DDM1. Science 2002; 297:1871–1873.
Kuijt SJ, Lamers GE, Rueb S, et al. Different subcellular localization and trafficking properties of KNOX class 1 homeodomain proteins from rice. Plant Mol Biol 2004; 55:781–796.
Xu J, Scheres B . Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 2005; 17:525–536.
Acknowledgements
We thank Veder Garcia (University of California, Berkeley, USA) for critically reading the paper, Zengyong He for providing the AtFKBP53::GUS transgenic line and Masami Horikoshi (The University of Tokyo, Japan) for the pET-6His-SpFkbp39P plasmid. This work was supported by grants from the National Science Foundation and US Department of Energy (to SL).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Li, H., Luan, S. AtFKBP53 is a histone chaperone required for repression of ribosomal RNA gene expression in Arabidopsis. Cell Res 20, 357–366 (2010). https://doi.org/10.1038/cr.2010.22
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2010.22
Keywords
This article is cited by
-
Chloroplast immunophilins
Protoplasma (2016)
-
An RNA-seq transcriptome analysis of floral buds of an interspecific Brassica hybrid between B. carinata and B. napus
Plant Reproduction (2014)
-
Proteins, the chaperone function and heredity
Biology & Philosophy (2013)
-
Genome-Wide Analysis of FK506-Binding Protein Genes in Populus trichocarpa
Plant Molecular Biology Reporter (2012)