Abstract
Autophagy is a bulk degradation system induced by cellular stresses such as nutrient starvation. Its function relies on the formation of double-membrane vesicles called autophagosomes. Unlike other organelles that appear to stably exist in the cell, autophagosomes are formed on demand, and once their formation is initiated, it proceeds surprisingly rapidly. How and where this dynamic autophagosome formation takes place has been a long-standing question, but the discovery of Atg proteins in the 1990's significantly accelerated our understanding of autophagosome biogenesis. In this review, we will briefly introduce each Atg functional unit in relation to autophagosome biogenesis, and then discuss the origin of the autophagosomal membrane with an introduction to selected recent studies addressing this problem.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Geng J, Baba M, Nair U, Klionsky DJ . Quantitative analysis of autophagy-related protein stoichiometry by fluorescence microscopy. J Cell Biol 2008; 182:129–140.
Mizushima N, Yamamoto A, Hatano M, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 2001; 152:657–668.
Fujita N, Hayashi-Nishino M, Fukumoto H, et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol Biol Cell 2008; 19:4651–4659.
Singh R, Cuervo AM . Autophagy in the cellular energetic balance. Cell Metab 2011; 13:495–504.
Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y . Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. J Cell Biol 1997; 139:1687–1695.
Mizushima N, Ohsumi Y, Yoshimori T . Autophagosome formation in mammalian cells. Cell Struct Funct 2002; 27:421–429.
Fujita N, Yoshimori T . Ubiquitination-mediated autophagy against invading bacteria. Curr Opin Cell Biol 2011; 23:492–497.
Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032–1036.
Komatsu M, Waguri S, Ueno T, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol 2005; 169:425–434.
Saitoh T, Fujita N, Jang MH, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008; 456:264–268.
Sou Y, Waguri S, Iwata J, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell 2008; 19:4762–4775.
Saitoh T, Fujita N, Hayashi T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA 2009; 106:20842–20846.
Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, Mizushima N . Autophagy is essential for preimplantation development of mouse embryos. Science 2008; 321:117–120.
Mizushima N, Komatsu M . Autophagy: renovation of cells and tissues. Cell 2011; 147:728–741.
Yang Z, Klionsky DJ . Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814–822.
Klionsky DJ, Cregg JM, Dunn WA Jr, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003; 5:539–545.
Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y . Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458–467.
Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y . The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20:5971–5981.
Suzuki K, Kubota Y, Sekito T, Ohsumi Y . Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells 2007; 12:209–218.
Itakura E, Mizushima N . Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764–776.
Hara T, Takamura A, Kishi C, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol 2008; 181:497–510.
Kabeya Y, Noda NN, Fujioka Y, Suzuki K, Inagaki F, Ohsumi Y . Characterization of the Atg17-Atg29-Atg31 complex specifically required for starvation-induced autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun 2009; 389:612–615.
Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y . Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell 2005; 16:2544–2553.
Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y . Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 2000; 150:1507–1513.
Chan EYW, Kir S, Tooze SA . siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J Biol Chem 2007; 282:25464–25474.
Young ARJ, Chan EYW, Hu XW, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888–3900.
Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y . Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell 2008; 19:2039–2050.
Cheong H, Nair U, Geng J, Klionsky DJ . The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2008; 19:668–681.
Chan EYW, Longatti A, McKnight NC, Tooze SA . Kinase-inactivated ULK proteins inhibit autophagy via their conserved C-terminal domains using an Atg13-independent mechanism. Mol Cell Biol 2009; 29:157–171.
Obara K, Noda T, Niimi K, Ohsumi Y . Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells 2008; 13:537–547.
Kihara A, Noda T, Ishihara N, Ohsumi Y . Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152:519–530.
Liang XH, Jackson S, Seaman M, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672–676.
Kihara A, Kabeya Y, Ohsumi Y, Yoshimori T . Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep 2001; 2:330–335.
Zhong Y, Wang QJ, Li X, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11:468–476.
Matsunaga K, Saitoh T, Tabata K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11:385–396.
Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q . Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA 2008; 105:19211–19216.
Itakura E, Kishi C, Inoue K, Mizushima N . Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360–5372.
Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ . The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6:79–90.
Lu Q, Yang P, Huang X, et al. The WD40 repeat PtdIns(3)P-binding protein EPG-6 regulates progression of omegasomes to autophagosomes. Dev Cell 2011; 21:343–357.
Orsi A, Razi M, Dooley HC, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 2012; 23:1860–1873.
Mizushima N, Noda T, Yoshimori T, et al. A protein conjugation system essential for autophagy. Nature 1998; 395:395–398.
Ishibashi K, Fujita N, Kanno E, et al. Atg16L2, a novel isoform of mammalian Atg16L that is not essential for canonical autophagy despite forming an Atg12-5-16L2 complex. Autophagy 2011; 7:1500–1513.
Fujita N, Saitoh T, Kageyama S, Akira S, Noda T, Yoshimori T . Differential involvement of Atg16L1 in Crohn disease and canonical autophagy: analysis of the organization of the Atg16L1 complex in fibroblasts. J Biol Chem 2009; 284:32602–32609.
Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F . Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem 2010; 285:1508–1515.
Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T . The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell 2008; 19:2092–2100.
Gammoh N, Florey O, Overholtzer M, Jiang X . Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol 2012; 20;144–149.
Nishimura T, Kaizuka T, Cadwell K, et al. FIP200 regulates targeting of Atg16L1 to the isolation membrane. EMBO Rep 2013; 14:284–291.
Fujita N, Morita E, Itoh T, et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J Cell Biol 2013; 203:115–128.
Romanov J, Walczak M, Ibiricu I, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J 2012; 31:4304–4317.
Hanada T, Noda NN, Satomi Y, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem 2007; 282:37298–37302.
Kirisako T, Ichimura Y, Okada H, et al. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J Cell Biol 2000; 151:263–276.
Ichimura Y, Kirisako T, Takao T, et al. A ubiquitin-like system mediates protein lipidation. Nature 2000; 408:488–492.
Kirisako T, Baba M, Ishihara N, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147:435–446.
Kabeya Y, Mizushima N, Ueno T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720–5728.
Nakatogawa H, Ichimura Y, Ohsumi Y . Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 2007; 130:165–178.
Itakura E, Kishi-Itakura C, Koyama-Honda I, Mizushima N . Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J Cell Sci 2012; 125:1488–1499.
Yamamoto H, Kakuta S, Watanabe TM, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol 2012; 198:219–233.
Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ . Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2010; 21:2257–2269.
Van der Vaart A, Griffith J, Reggiori F . Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell 2010; 21:2270–2284.
Ohashi Y, Munro S . Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell 2010; 21:3998–4008.
Yen W-L, Shintani T, Nair U, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol 2010; 188:101–114.
Shirahama-Noda K, Kira S, Yoshimori T, Noda T . TRAPPIII is responsible for the vesicular transport from early endosomes to the Golgi apparatus that facilitates Atg9 cycling in autophagy. J Cell Sci 2013; 126(Pt 21):4963–4973.
Axe EL, Walker SA, Manifava M, et al. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685–701.
Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A . A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol 2009; 11:1433–1437.
Ylä-Anttila P, Vihinen H, Jokitalo E, Eskelinen E-L . 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009; 5:1180–1185.
Matsunaga K, Morita E, Saitoh T, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 2010; 190:511–521.
Fan W, Nassiri A, Zhong Q . Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci USA 2011; 108:7769–7774.
Hamasaki M, Furuta N, Matsuda A, et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013; 495:389–393.
Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N . Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 2013; 9:1491–1499.
Bodemann BO, Orvedahl A, Cheng T, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 2011; 144:253–267.
Karanasios E, Stapleton E, Manifava M, et al. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci 2013 Sep 6. doi:10.1242/jcs.132415
Tian Y, Li Z, Hu W, et al. C. elegans screen identifies autophagy genes specific to multicellular organisms. Cell 2010; 141:1042–1055.
Hailey DW, Rambold AS, Satpute-Krishnan P, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141:656–667.
Rowland AA, Voeltz GK . Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol 2012; 13:607–625.
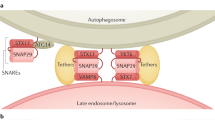
Itakura E, Kishi-Itakura C, Mizushima N . The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012; 151:1256–1269.
Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y . Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci 2013; 126:2534–2544.
Graef M, Friedman JR, Graham C, Babu M, Nunnari J . ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell 2013; 24:2918–2931.
Hamasaki M, Noda T, Ohsumi Y . The early secretory pathway contributes to autophagy in yeast. Cell Struct Funct 2003; 28:49–54.
Ishihara N, Hamasaki M, Yokota S, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell 2001; 12:3690–3702.
Guo Y, Chang C, Huang R, Liu B, Bao L, Liu W . AP1 is essential for generation of autophagosomes from the trans-Golgi network. J Cell Sci 2012; 125:1706–1715.
Zoppino FCM, Militello RD, Slavin I, Alvarez C, Colombo MI . Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 2010; 11:1246–1261.
Ge L, Melville D, Zhang M, Schekman R . The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife 2013; 2:e00947.
Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC . Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol 2010; 12:747–757.
Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC . Autophagosome precursor maturation requires homotypic fusion. Cell 2011; 146:303–317.
Kageyama S, Omori H, Saitoh T, et al. The LC3 recruitment mechanism is separate from Atg9L1-dependent membrane formation in the autophagic response against Salmonella. Mol Biol Cell 2011; 22:2290–2300.
Longatti A, Lamb CA, Razi M, Yoshimura S-I, Barr FA, Tooze SA . TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 2012; 197:659–675.
Knaevelsrud H, Søreng K, Raiborg C, et al. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. J Cell Biol 2013; 202:331–349.
Puri C, Renna M, Bento CF, Moreau K, Rubinsztein DC . Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell 2013; 154:1285–1299.
Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F . An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 2010; 190:1005–1022.
Lynch-Day MA, Bhandari D, Menon S, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci USA 2010; 107:7811–7816.
Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y . Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J Biol Chem 2012; 287:44261–44269.
Nair U, Jotwani A, Geng J, et al. SNARE proteins are required for macroautophagy. Cell 2011; 146:290–302.
Ragusa MJ, Stanley RE, Hurley JH . Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell 2012; 151:1501–1512.
Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y . Atg17 recruits Atg9 to organize the pre-autophagosomal structure. Genes Cells 2009; 14:525–538.
Youle RJ, Narendra DP . Mechanisms of mitophagy. Nat Rev Mol Cell Biol 2011; 12:9–14.
Huett A, Heath RJ, Begun J, et al. The LRR and RING domain protein LRSAM1 is an E3 ligase crucial for ubiquitin-dependent autophagy of intracellular Salmonella typhimurium. Cell Host Microbe 2012; 12:778–790.
Manzanillo PS, Ayres JS, Watson RO, et al. The ubiquitin ligase parkin mediates resistance to intracellular pathogens. Nature 2013; 501:512–516.
Kim PK, Hailey DW, Mullen RT, Lippincott-Schwartz J . Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc Natl Acad Sci USA 2008; 105:20567–20574.
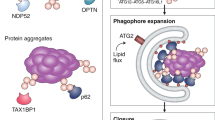
Kirkin V, Lamark T, Sou YS, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell 2009; 33:505–516.
Pankiv S, Clausen TH, Lamark T, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131–24145.
Wild P, Farhan H, McEwan DG, et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011; 333:228–233.
Thurston TLM, Ryzhakov G, Bloor S, von Muhlinen N, Randow F . The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat Immunol 2009; 10:1215–1221.
Huang J, Birmingham CL, Shahnazari S, et al. Antibacterial autophagy occurs at PI(3)P-enriched domains of the endoplasmic reticulum and requires Rab1 GTPase. Autophagy 2011; 7:17–26.
Maejima I, Takahashi A, Omori H, et al. Autophagy sequesters damaged lysosomes to control lysosomal biogenesis and kidney injury. EMBO J 2013; 32:2336–2347.
Hung YH, Chen LM, Yang JY, Yang WY . Spatiotemporally controlled induction of autophagy-mediated lysosome turnover. Nat Commun 2013; 4:2111.
Acknowledgements
We thank our lab member Marija Landekic for discussion and English proofreading. Work in the authors' lab is supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and by Japan Science and Technology Agency CREST.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shibutani, S., Yoshimori, T. A current perspective of autophagosome biogenesis. Cell Res 24, 58–68 (2014). https://doi.org/10.1038/cr.2013.159
Published:
Issue date:
DOI: https://doi.org/10.1038/cr.2013.159
Keywords
This article is cited by
-
Transcription factors RhPIF4/8 and RhHY5 regulate autophagy-mediated petal senescence in rose (Rosa hybrida)
Horticulture Advances (2023)
-
Morphine-induced microglial immunosuppression via activation of insufficient mitophagy regulated by NLRX1
Journal of Neuroinflammation (2022)
-
The dual role of autophagy in acute myeloid leukemia
Journal of Hematology & Oncology (2022)
-
Dendritic autophagy degrades postsynaptic proteins and is required for long-term synaptic depression in mice
Nature Communications (2022)
-
Is targeting autophagy mechanism in cancer a good approach? The possible double-edge sword effect
Cell & Bioscience (2021)