- TECHNOLOGY FEATURE
Cellular censuses to guide cancer care
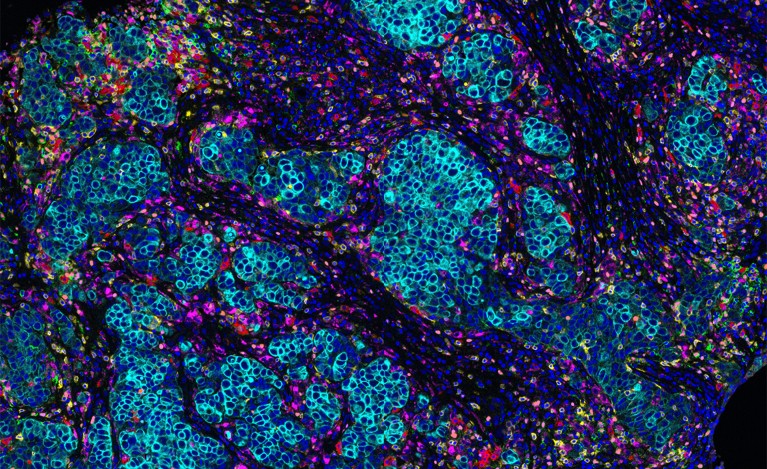
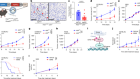
The microenvironment surrounding a breast-cancer tumour (blue) contains different types of immune cells, such as T cells and B cells. Credit: Akoya Biosystems
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Rent or buy this article
Prices vary by article type
from$1.95
to$39.95
Prices may be subject to local taxes which are calculated during checkout
Nature 567, 555-557 (2019)
doi: https://doi.org/10.1038/d41586-019-00904-5
References
Keren, L. et al. Cell 174, 1373–1387 (2018).
Lavin, Y. et al. Cell 169, 750–765 (2017).
Jerby-Arnon, L. et al. Cell 175, 984–997 (2018).
Giesen, C. et al. Nature Meth. 11, 417–422 (2014).
Angelo, M. et al. Nature Med. 20, 436–442 (2014).
Salmen, F. et al. Preprint at bioRxiv https://doi.org/10.1101/358937 (2018).
Chevrier, S. et al. Cell 169, 736–749 (2017).
Thorsson, V. et al. Immunity 48, 812–830 (2018).
Saltz, J. et al. Cell Rep. 23, 181–193 (2018).

 How to build a human cell atlas
How to build a human cell atlas
 Single-cell approaches to immune profiling
Single-cell approaches to immune profiling
 NatureTech hub
NatureTech hub