Molecular glue degraders (MGDs) are a powerful approach to target proteins traditionally considered ‘undruggable’. By harnessing the ubiquitin–proteasome system, they can induce protein degradation even in the absence of conventional binding sites. However, their discovery has often relied on serendipity, leaving much of their potential untapped.
Lacking rational design strategies, researchers have turned to phenotypic methods such as cell-viability assays; but these approaches may miss therapeutically relevant targets that do not affect viability.
Zoran Rankovic, director of the Centre for Protein Degradation at the Institute of Cancer Research (ICR), London, explained: “The traditional approach of screening thousands of compounds across multiple cell lines is time-consuming and often uncovers only a limited number of targets. Promising candidates are frequently missed—either because they are not essential or because the compound lacks sufficient potency.” Rankovic likened these conventional discovery methods to ‘panning for gold’—resource-intensive, heavily reliant on chance, and with low success rates. “In contrast, NEOsphere Biotechnologies’ high-throughput proteomics is like putting on goggles that let you see the gold nuggets and pick them up easily—it’s eye-opening.”
A data-driven paradigm shift
NEOsphere Biotechnologies’ integrated high-throughput proteomics platform enables rapid screening of entire degrader libraries and systematic identification of compounds targeting proteins of interest.
For Rankovic, this represents a fundamental shift in how his lab approaches MGD drug discovery. “Together with NEOsphere Biotechnologies, ICR recently began screening a 5,000-compound molecular glue library, and within just a few months, we’ve already identified several potent and selective degraders for clinically relevant targets previously considered out of reach,” he explained.
Proteomics data also guide hit optimization. “We use it to support structure–activity relationship (SAR) studies, which are particularly valuable for molecular glues due to their often steep and unpredictable SAR profiles. The ability to rapidly assess proteome-wide effects of compound modifications enables faster iteration and more-efficient refinement of promising candidates. As we initiate our first lead-optimization programs and broaden proteomic screening across the full library, we aim to advance the most promising leads to a stage where they can be offered as licensing-ready programs to pharmaceutical partners.”
MGDs are small molecules that redirect E3 ligases to selectively degrade proteins. Their therapeutic potential has been demonstrated by drugs such as Revlimid (lenalidomide), which improves multiple myeloma outcomes. However, discovery beyond serendipity remains challenging due to complex SARs and the limited sensitivity of traditional methods.
Rankovic was initially skeptical about using proteomics to address these challenges. Since it had long been a bottleneck in drug discovery, applying proteomics in a high-throughput format to screen entire libraries seemed implausible. However, a pilot study during his tenure at St. Jude Children’s Research Hospital in Memphis quickly dispelled these doubts.
In just a few weeks—the time it would normally take Rankovic to receive proteomics data for only a handful of compounds—NEOsphere Biotechnologies provided extensive data for 100 cereblon (CRBN) ligands across two cell lines, identifying nearly 50 previously unreported neosubstrates. Published in Nature Communications, this study greatly expanded the known CRBN neosubstrate space and detected even weakly degraded targets—a key breakthrough in the field1. Importantly, it also identified neosubstrates lacking classical CRBN-degron motifs, including the stress granule protein Ras GTPase-activating protein-binding protein 2 (G3BP2) and the lysine-specific demethylase 4B (KDM4B), highlighting the platform’s power to reveal novel targets even without a full mechanistic understanding of MGD-mediated degradation. These findings indicate that the field has only just begun to uncover the full range of proteins amenable to targeted degradation.
The quality of the results also supported follow-up chemistry optimization. After discovering a degrader for G3BP2—a protein with no known inhibitors—researchers at St. Jude used the proteomics platform to guide the synthesis of analogs, an approach that ultimately resulted in a more potent G3BP2 degrader.
The success of the pilot study led Rankovic to continue the collaboration at ICR. “We now routinely analyze over 11,000 proteins per sample—well beyond the typical depth of most proteomics studies, which usually cover only around 9,000 to 10,000 proteins. This gives us a much more comprehensive view and unprecedented insight into how small molecules interact with the proteome as a whole.”
Accelerating degrader drug discovery
NEOsphere Biotechnologies accelerates degrader discovery across all stages—from hit identification to lead optimization and toxicity assessment. Its fully automated, high-throughput proteomics platform combines label-free, data-independent acquisition (DIA) mass spectrometry with advanced data analysis, offering proteome-wide insights with exceptional data quality and completeness in disease-relevant contexts (Fig. 1).
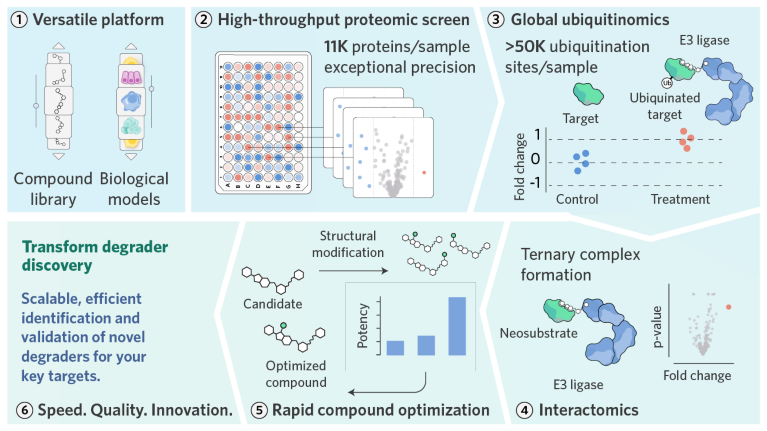
Fig. 1 | NEOsphere Biotechnologies’ integrated high-throughput proteomics platform accelerates drug discovery from hit identification to lead optimization. The platform screens all types of protein degraders and stabilizers across cell lines, tissues, and patient-derived material to systematically identify compounds targeting disease-relevant proteins. Integrated global ubiquitinomics and interactomics reveal compound-induced ubiquitination and ternary complex formation, while proteome-wide data on structure–activity relationships, selectivity, and potency drive rapid lead optimization.
The company analyzes all types of protein degraders, stabilizers, and proximity-inducing compounds—including MGDs, proteolysis-targeting chimeras (PROTACs), monovalent degraders, deubiquitinase inhibitors, and degrader–antibody conjugates—and is compatible with a wide range of biological materials, from cell lines and primary cells to patient-derived samples and tissues.
Library screening and degrader optimization
Novel degraders for relevant targets can be identified and validated using NEOsphere Biotechnologies’ unique proteomics portfolio. High-throughput global proteomics rapidly profiles tens of thousands of compounds in a native, ligase-independent context, enabling unbiased target discovery under physiologically relevant conditions. Optimized for coverage and precision, it reliably detects low-abundance targets and subtle regulatory effects, generating leads for further development. Mapping each compound’s full target spectrum and monitoring proteome-wide downregulation transforms a once serendipitous process into a systematic, compound-centric approach to drug discovery.
Complementing this, high-throughput interactomics reveals ternary complex formation between compounds, E3 ligases, and target proteins, providing mechanistic insight into degrader activity. Identified hits can then be refined through global proteomics, where proteome-wide data on selectivity, potency, efficacy, and degradation kinetics guide iterative medicinal chemistry.
“Global proteomics shows us which compounds cause protein downregulation, while interactomics reveals compound-induced proximity between E3 ligases and target proteins,” explained Henrik Daub, CSO at NEOsphere Biotechnologies. This mechanistic insight is particularly valuable early in the discovery process. “Some compounds bind targets but aren’t yet optimized to trigger degradation. Interactomics identifies these early interactions, helping us prioritize compounds for further development.”
By combining these approaches, researchers gain a fuller, more nuanced understanding of degrader function inside cells. “Both methods tackle the same fundamental challenge: finding the best starting points for drug discovery,” Daub concluded. “Integrating these data provides clearer insight into which molecular glues hold real therapeutic promise—and why.”
Unlocking the global ubiquitinome
Once promising MGD candidates are identified, validation becomes critical. Demonstrating that a compound induces target ubiquitination in addition to ternary complex formation and protein downregulation provides strong evidence of direct degradation. To this end, NEOsphere Biotechnologies’ global ubiquitinomics offers detailed insights into both target-specific and system-wide ubiquitination events. By quantifying up to 60,000 ubiquitination sites per sample, it generates high-resolution data on how degrader compounds rapidly modulate cellular ubiquitination.
Daub explained how this clarifies target engagement: “A consistent increase in specific ubiquitination sites within minutes of compound treatment—especially when aligned with target downregulation observed in global proteomics—is a strong indicator of direct degrader activity.” Many partners now routinely use global ubiquitinomics as a follow-up to proteomic screening or to validate the mechanism of action for advanced compounds.
“Global ubiquitinomics has become an essential step,” Daub added. “It confirms compound-induced target ubiquitination, giving confidence to advance the right compounds and targets. When global proteomics, ubiquitinomics, and interactomics all point in the same direction,” Daub concluded, “you’re not just observing an effect—you’re uncovering the mechanism of action of your compound. And that understanding initiates and drives rational degrader development.”
Translating proteomics data into actionable insights
As mass spectrometry technology advances and sample throughput grows, scalable, parallelizable, fully automated methods for analysis and interpretation of large-scale, high-dimensional data are increasingly critical to harness proteomic insights in drug discovery.
To meet this need, NEOsphere Biotechnologies developed neoVERSE—user-friendly software for the seamless analysis of extensive proteomics data. Combining advanced visualization with robust statistical and meta-analysis tools, neoVERSE makes complex data accessible to a wide range of users. It ensures rigorous quality control, and enables researchers to evaluate compound performance both at the individual sample level and across entire projects (Fig. 2).
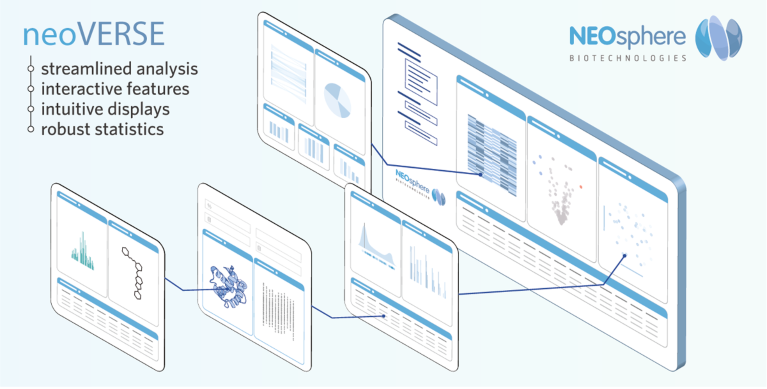
Fig. 2 | neoVERSE intuitive software streamlines the analysis of large-scale proteomics data. With advanced visualizations and statistical analyses, it enables seamless data review and project evaluation, turning complex datasets into usable insights.
“We created neoVERSE to bridge the gap between complex data and fast, confident decision-making,” said Daub. With intuitive interfaces, researchers can navigate datasets with ease and quickly extract relevant insights. “Potent and selective compounds can be identified, their efficacy evaluated, and proteomic results seamlessly integrated into discovery programs—all within just a few clicks,” Daub added. “In short, neoVERSE turns complex proteomic data into actionable insights that accelerate informed drug discovery.”
From trial-and-error to systematic scalability
NEOsphere Biotechnologies is transforming degrader drug discovery, turning a traditionally slow and serendipitous process into a systematic, scalable approach. Its integrated proteomics platform allows researchers to map the full target landscape of their degraders and unlock the potential of previously undruggable proteins. Optimized for precision, efficiency, and speed, the platform accelerates rational degrader discovery—translating cutting-edge science into real-world therapeutic impact.


