Abstract
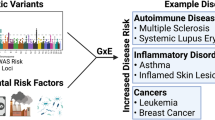
Genetic and environmental risk factors and their interactions contribute to the development of complex diseases. In this review, we discuss methodological issues involved in investigating gene–environment (G × E) interactions in genetic–epidemiological studies of complex diseases and their potential relevance for clinical application. Although there are some important examples of interactions and applications, the widespread use of the knowledge about G × E interaction for targeted intervention or personalized treatment (pharmacogenetics) is still beyond current means. This is due to the fact that convincing evidence and high predictive or discriminative power are necessary conditions for usefulness in clinical practice. We attempt to clarify conceptual differences of the term ‘interaction’ in the statistical and biological sciences, since precise definitions are important for the interpretation of results. We argue that the investigation of G × E interactions is more rewarding for the detailed characterization of identified disease genes (ie at advanced stages of genetic research) and the stratified analysis of environmental effects by genotype or vice versa. Advantages and disadvantages of different epidemiological study designs are given and sample size requirements are exemplified. These issues as well as a critical appraisal of common methodological concerns are finally discussed.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Ordovas JM, Mooser V : Nutrigenomics and nutrigenetics. Curr Opin Lipidol 2004; 15: 101–108.
Brennan P : Gene–environment interaction and aetiology of cancer: what does it mean and how can we measure it? Carcinogenesis 2002; 23: 381–387.
Gardiner SJ, Begg EJ : Pharmacogenetic testing for drug metabolizing enzymes: is it happening in practice? Pharmacogenet Genomics 2005; 15: 365–369.
Rees JL : The genetics of sun sensitivity in humans. Am J Hum Genet 2004; 75: 739–751.
Hauser ER, Allen AS : Where the rubber meets the road in pharmacogenetics: assessment of gene–environment interactions. Am Heart J 2003; 146: 929–931.
Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG : Replication validity of genetic association studies. Nat Genet 2001; 29: 306–309.
Rose G : Sick individuals and sick populations. Int J Epidemiol 2001; 30: 427–432.
Willett WC : Balancing life-style and genomics research for disease prevention. Science 2002; 296: 695–698.
Roses AD : Pharmacogenetics and drug development: the path to safer and more effective drugs. Nat Rev Genet 2004; 5: 645–656.
Hillman MA, Wilke RA, Yale SH et al: A prospective, randomized pilot trial of model-based warfarin dose initiation using CYP2C9 genotype and clinical data. Clin Med Res 2005; 3: 137–145.
Voora D, Eby C, Linder MW et al: Prospective dosing of warfarin based on cytochrome P-450 2C9 genotype. Thromb Haemost 2005; 93: 700–705.
Goldstein DB, Tate SK, Sisodiya SM : Pharmacogenetics goes genomic. Nat Rev Genet 2003; 4: 937–947.
Cordell HJ : Epistasis: what it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum Mol Genet 2002; 11: 2463–2468.
Rothman KJ, Greenland S : Modern Epidemiology. Philadelphia: Lippincott-Raven, 1998.
Yang Q, Khoury MJ : Evolving methods in genetic epidemiology. III. Gene–environment interaction in epidemiologic research. Epidemiol Rev 1997; 19: 33–43.
Ottman R : An epidemiologic approach to gene–environment interaction. Genet Epidemiol 1990; 7: 177–185.
Thomas DC : Temporal effects and interactions in cancer: implications of carcinogenic models; in Prentice RL, Whittemore AS (eds): Environmental Epidemiology: Risk Assessment. Philadelphia: Society for Industrial and Applied Mathematics, 1982, pp 107–121.
Yusuf S, Wittes J, Probstfield J, Tyroler HA : Analysis and interpretation of treatment effects in subgroups of patients in randomized clinical trials. JAMA 1991; 266: 93–98.
Tukey JW : One degree of freedom for non-additivity. Biometrics 1949; 5: 232–242.
Thompson WD : Effect modification and the limits of biological inference from epidemiologic data. J Clin Epidemiol 1991; 44: 221–232.
Clayton D, McKeigue PM : Epidemiological methods for studying genes and environmental factors in complex diseases. Lancet 2001; 358: 1356–1360.
Davey Smith G, Ebrahim S : ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003; 32: 1–22.
Gauderman WJ, Thomas DC : The role of interacting determinants in the localization of genes. Adv Genet 2001; 42: 393–412.
Dizier MH, Selinger-Leneman H, Genin E : Testing linkage and gene × environment interaction: comparison of different affected sib-pair methods. Genet Epidemiol 2003; 25: 73–79.
Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ : Exploiting gene–environment interaction to detect genetic associations. Hum Hered 2007; 63: 111–119.
Schaid DJ : Case–parents design for gene–environment interaction. Genet Epidemiol 1999; 16: 261–273.
Witte JS, Gauderman WJ, Thomas DC : Asymptotic bias and efficiency in case–control studies of candidate genes and gene–environment interactions: basic family designs. Am J Epidemiol 1999; 149: 693–705.
Gauderman WJ, Witte JS, Thomas DC : Family-based association studies. J Natl Cancer Inst Monogr 1999; 26: 31–37.
Hunter DJ : Gene–environment interactions in human diseases. Nat Rev Genet 2005; 6: 287–298.
Wacholder S, Garcia-Closas M, Rothman N : Study of genes and environmental factors in complex diseases. Lancet 2002; 359: 1155.
Burton P, McCarthy M, Elliott P : Study of genes and environmental factors in complex diseases. Lancet 2002; 359: 1155–1156.
Stene LC : Study of genes and environmental factors in complex diseases. Lancet 2002; 359: 1156.
Banks E, Meade T : Study of genes and environmental factors in complex diseases. Lancet 2002; 359: 1156–1157.
Khoury MJ, Flanders WD : Nontraditional epidemiologic approaches in the analysis of gene–environment interaction: case–control studies with no controls!. Am J Epidemiol 1996; 144: 207–213.
Schmidt S, Schaid DJ : Potential misinterpretation of the case-only study to assess gene–environment interaction. Am J Epidemiol 1999; 150: 878–885.
Gatto NM, Campbell UB, Rundle AG, Ahsan H : Further development of the case-only design for assessing gene–environment interaction: evaluation of and adjustment for bias. Int J Epidemiol 2004; 33: 1014–1024.
Garcia-Closas M, Thompson WD, Robins JM : Differential misclassification and the assessment of gene–environment interactions in case–control studies. Am J Epidemiol 1998; 147: 426–433.
Vineis P : A self-fulfilling prophecy: are we underestimating the role of the environment in gene–environment interaction research? Int J Epidemiol 2004; 33: 945–946.
Umbach DM, Weinberg CR : Designing and analysing case–control studies to exploit independence of genotype and exposure. Stat Med 1997; 16: 1731–1743.
Liu X, Fallin MD, Kao WH : Genetic dissection methods: designs used for tests of gene–environment interaction. Curr Opin Genet Dev 2004; 14: 241–245.
Chatterjee N, Kalaylioglu Z, Carroll RJ : Exploiting gene–environment independence in family-based case–control studies: increased power for detecting associations, interactions and joint effects. Genet Epidemiol 2005; 28: 138–156.
Abel L, Dessein AJ : Genetic epidemiology of infectious diseases in humans: design of population-based studies. Emerg Infect Dis 1998; 4: 593–603.
Hill AV : Genetics and genomics of infectious disease susceptibility. Br Med Bull 1999; 55: 401–413.
Clementi M, Di Gianantonio E : Genetic susceptibility to infectious diseases. Reprod Toxicol 2006; 21: 345–349.
Smith MW, Dean M, Carrington M et al: Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC), ALIVE Study. Science 1997; 277: 959–965.
Pouniotis DS, Proudfoot O, Minigo G, Hanley JL, Plebanski M : Malaria parasite interactions with the human host. J Postgrad Med 2004; 50: 30–34.
Brown P, Cervenakova L, Goldfarb LG et al: Iatrogenic Creutzfeldt–Jakob disease: an example of the interplay between ancient genes and modern medicine. Neurology 1994; 44: 291–293.
Cardon LR, Idury RM, Harris TJ, Witte JS, Elston RC : Testing drug response in the presence of genetic information: sampling issues for clinical trials. Pharmacogenetics 2000; 10: 503–510.
Kelly PJ, Stallard N, Whittaker JC : Statistical design and analysis of pharmacogenetic trials. Stat Med 2005; 24: 1495–1508.
Hein R, Beckmann L, Chang-Claude J : Sample size requirements for indirect association studies of gene–environment interactions (G × E). Genet Epidemiol 2008; 32: 235–245.
Riboli E, Hunt KJ, Slimani N et al: European Prospective Investigation into Cancer and Nutrition (EPIC): study populations and data collection. Public Health Nutr 2002; 5: 1113–1124.
Kolonel LN, Altshuler D, Henderson BE : The multiethnic cohort study: exploring genes, lifestyle and cancer risk. Nat Rev Cancer 2004; 4: 519–527.
Luan JA, Wong MY, Day NE, Wareham NJ : Sample size determination for studies of gene–environment interaction. Int J Epidemiol 2001; 30: 1035–1040.
Foppa I, Spiegelman D : Power and sample size calculations for case–control studies of gene–environment interactions with a polytomous exposure variable. Am J Epidemiol 1997; 146: 596–604.
Judson R : Using multiple drug exposure levels to optimize power in pharmacogenetic trials. J Clin Pharmacol 2003; 43: 816–824.
Singer C, Grossman I, Avidan N, Beckmann JS, Pe’er I : Trick or treat: the effect of placebo on the power of pharmacogenetic association studies. Hum Genomics 2005; 2: 28–38.
Elston RC, Idury RM, Cardon LR, Lichter JB : The study of candidate genes in drug trials: sample size considerations. Stat Med 1999; 18: 741–751.
Garcia-Closas M, Lubin JH : Power and sample size calculations in case–control studies of gene–environment interactions: comments on different approaches. Am J Epidemiol 1999; 149: 689–692.
Gauderman WJ : Sample size requirements for matched case–control studies of gene–environment interaction. Stat Med 2002; 21: 35–50.
Saunders CL, Bishop DT, Barrett JH : Sample size calculations for main effects and interactions in case–control studies using Stata's nchi2 and npnchi2 functions. Stata J 2003; 3: 47–56.
Ryan SG : Regression to the truth: replication of association in pharmacogenetic studies. Pharmacogenomics 2003; 4: 201–207.
Acknowledgements
This work was funded by the Bundesministerium für Bildung und Forschung through the German National Genome Net (NGFN2, grant numbers 01GR0460 and 01GR0461).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dempfle, A., Scherag, A., Hein, R. et al. Gene–environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet 16, 1164–1172 (2008). https://doi.org/10.1038/ejhg.2008.106
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2008.106
Keywords
This article is cited by
-
Genome-wide case-only analysis of gene-gene interactions with known Parkinson’s disease risk variants reveals link between LRRK2 and SYT10
npj Parkinson's Disease (2023)
-
Genotype imputation in case-only studies of gene-environment interaction: validity and power
Human Genetics (2021)
-
Interaction between lifestyle behaviors and genetic polymorphism in SCAP gene on blood pressure among Chinese children
Pediatric Research (2019)
-
Childhood adversity and parenting behavior: the role of oxytocin receptor gene polymorphisms
Journal of Neural Transmission (2019)
-
Interactions between breast cancer susceptibility loci and menopausal hormone therapy in relationship to breast cancer in the Breast and Prostate Cancer Cohort Consortium
Breast Cancer Research and Treatment (2016)