Abstract
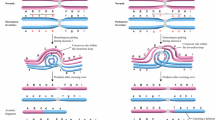
Paracentric inversions (PAIs) are structural chromosomal rearrangements generally considered to be harmless. To date, only a few studies have been performed concerning the meiotic segregation of these rearrangements, using either the human–hamster fertilization system or fluorescence in situ hybridization (FISH) with centromeric or telomeric DNA probes. To improve the assessment of imbalances in PAI, we present a new strategy based on FISH assay using multiple bacterial artificial chromosome probes, which allow a precise localization of chromosome break points and the identification of all meiotic products in human sperm. Sperm samples of three cases with PAI were investigated: an inv(5)(q13.2q33.1), an inv(9)(q21.2q34.13) and an inv(14)(q23.2q32.13). The frequencies of spermatozoa with inverted chromosomes were 44.7% in inv(5), 42.7% in inv(9) and 46.7% in inv(14). The global incidences of unbalanced complements were 9.7, 12.6 and 3.7% in inv(5), inv(9) and inv(14), respectively. This report is the first study providing a detailed description of meiotic segregation patterns in human sperm by using a sperm FISH approach. This study demonstrates that the detailed analysis of segregation in PAI may provide important data for both genetic analysis and counseling of inversion carriers.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Madan K : Paracentric inversions: a review. Hum Genet 1995; 96: 503–515.
Fryns J, Kleczkowska A, kubien E, van den Berghe H : Structural chromosome rearrangements in couples with recurrent fetal wastage: the leuven experience; in Sandberg A (ed): The Cytogenetics of Mammalian Autosomal Rearrangements. New York: Alan R Liss, 1988, Vol 8, pp 453–470.
Worsham MJ, Miller DA, Devries JM et al: A dicentric recombinant 9 derived from a paracentric inversion: phenotype, cytogenetics, and molecular analysis of centromeres. Am J Hum Genet 1989; 44: 115–123.
Mules EH, Stamberg J : Reproductive outcomes of paracentric inversion carriers; report of a liveborn dicentric recombinant and literature review. Hum Genet 1984; 67: 126–131.
Whiteford ML, Baird C, Kinmond S, Donaldson B, Davidson HR : A child with bisatellited, dicentric chromosome 15 arising from a maternal paracentric inversion of chromosome 15q. J Med Genet 2000; 37: E11.
Lefort G, Blanchet P, Belgrade N et al: Stable dicentric duplication-deficiency chromosome 14 resulting from crossing-over within a maternal paracentric inversion. Am J Hum Genet 2002; 113: 333–338.
Phelan MC, Stevenson RE, Anderson Jr EV : Recombinant chromosome 9 possibly derived from breakage and reunion of sister chromatids within a paracentric inversion loop. Am J Med Genet 1993; 46: 304–308.
Yang SP, Bidichandani SI, Figuera LE et al: Molecular analysis of deletion (17)(p11.2p11.2) in a family segregating a 17p paracentric inversion: implications for carriers of paracentric inversions. Am J Hum Genet 1997; 60: 1184–1193.
Feldman GL, Weiss L, Phelan M, Shroer RJ, Van Dyke DL : Inverted duplication of 8p: ten new patients and review of the literature. Am J Med Genet 1993; 47: 482–486.
Mitchell JJ, Vekemans M, Luscombe S et al: U-type exchange in a paracentric inversion as a possible mechanism of origin of an inverted tandem duplication of chromosome 8. Am J Med Genet 1994; 49: 384–387.
Pettenati MJ, Rao PN, Phelan MC et al: Paracentric inversions in humans: a review of 446 paracentric inversions with presentation of 120 new cases. Am J Med Genet 1995; 55: 171–187.
Sutherland GR, Callen DF, Gardner RJM : Paracentric inversions do not normally generate monocentric recombinant chromosomes. Am J Med Genet 1995; 59: 390.
Madan K, Nieuwint AW : Reproductive risks for paracentric inversion heterozygotes: inversion or insertion? That is the question. Am J Med Genet 2002; 107: 340–343.
Cheng EY, Chen YJ, Disteche CM, Gartler SM : Analysis of a paracentric inversion in human oocytes: nonhomologous pairing in pachytene. Hum Genet 1999; 105: 191–196.
Brown GM, Leversha M, Hulten M, Ferguson-Smith MA, Affara NA, Furlong RA : Genetic analysis of meiotic recombination in humans by use of sperm typing: reduced recombination within a heterozygous paracentric inversion of chromosome 9q32–q34.3. Am J Hum Genet 1998; 62: 1484–1492.
Pellestor F, Andréo B, Puechberty J, Lefort G, Sarda P : Analysis of sperm aneuploidy by PRINS; in Pellestor F (ed): PRINS and in situ PCR protocols, second edn. Totowa: Humana Press, 2006, pp 49–60.
Bhatt S, Moradkhani K, Mrasek K et al: Breakpoint characterization: a new approach for segregation analysis of paracentric inversion in human sperm. Mol Hum Reprod 2007; 13: 751–756.
Martin RH : Sperm chromosome analysis in a man heterozygous for a paracentric inversion of chromosome 7 (q11q22). Hum Genet 1986; 73: 97–100.
Martin RH : Sperm chromosome analysis in a man heterozygous for a paracentric inversion of chromosome 14 (q24.1q32.1). Am J Hum Genet 1999; 64: 1480–1484.
Devine DH, Whitman-Elia G, Best RG, Edwards JG : Paternal paracentric inversion of chromosome 2: a possible association with recurrent pregnancy loss and infertility. J Assist Reprod Genet 2000; 17: 293–296.
Anton E, Vidal F, Egozcue J, Blanco J : Genetic reproductive risk in inversion carriers. Fertil Steril 2006; 85: 661–666.
Vialard F, Delanete A, Clement P, Simon-Bouy B, Aubriot FX, Selva J : Sperm chromosome analysis in two cases of paracentric inversion. Fertil Steril 2007; 87: 418. e1–418. e5.
Anton E, Blanco J, Egozcue J, Vidal F : Risk assessment and segregation analysis in a pericentric inversion inv6p23q25 carrier using FISH on decondensed sperm nuclei. Cytogenet Genome Res 2002; 97: 149–154.
Mikhaail-Philips MM, McGillivray BC, Hamilton SJ et al: Unusual segregation products in sperm from a pericentric inversion 17 heterozygote. Hum Genet 2005; 117: 357–365.
Chantot-Bastaraud S, Ravel C, Berthaut I et al: Sperm-FISH analysis in a pericentric chromosome 1 inversion, 46, XY, inv (1) (p22q42), associated with infertility. Mol Hum Reprod 2007; 13: 55–59.
Anton E, Blanco J, Egozcue J, Vidal F : Sperm studies in heterozygote inversion carriers: a review. Cytogenet Genome Res 2005; 111: 297–304.
Morel F, Laudier B, Guerif F et al: Meiotic segregation analysis in spermatozoa of pericentric inversion carriers using fluorescence in situ hybridization. Hum Reprod 2007; 22: 136–141.
Jaarola M, Martin Rh, Ashley T : Direct evidence for suppression of recombination within two pericentric inversions in humans: a new sperm FISH technique. Am J Hum Genet 1998; 63: 218–224.
Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B : A high-resolution recombination map of the human genome. Nat Genet 2002; 31: 241–247.
Sun F, Oliver-Bonet M, Liehr T et al: Human male recombination maps for individual chromosomes. Am J Hum Genet 2004; 74: 521–531.
Ciccone R, Mattina T, Giorda R et al: Inversion polymorphisms and non-contiguous terminal deletions: the cause and the (unpredicted) effect of our genome architecture. J Med Genet 2006; 43: e19.
South ST, Swensen JJ, Maxwell T, Rope A, Brothman AR, Chen Z : A new genomic mechanism leading to cri-du-chat syndrome. Am J Med Genet A 2006; 140: 2714–2720.
Ashley T : G-band position effects on meiotic synapsis and crossing over. Genetics 1988; 118: 307–317.
Manvelyan M, Shreyer I, Hols-Herpertz I et al: Forty-eight cases with infertility due to balanced chromosomal rearrangements: detailed molecular cytogenetic analysis of the 90 involved breakpoints. Int J Mol Med 2006; 19: 855–864.
Stankiewicz P, Lupski JR : Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 2002; 12: 312–319.
Giorda R, Ciccone R, Gimelli G et al: Two classes of low-copy repeats comediate a new recurrent rearrangement consisting of duplication at 8p23.1 and triplication at 8p23.2. Hum Mutat 2007; 28: 459–468.
Flores M, Morales L, Gonzaga-Jauregui C et al: Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci USA 2007; 104: 6099–6106.
Feuk L, Carson AR, Scherer SW : Structural variation in the human genome. Nat Rev Genet 2006; 7: 85–97.
Koehler KE, Millie EA, Cherry JP et al: Sex-specific differences in meiotic chromosome segregation revealed by dicentric bridge resolution in mice. Genetics 2002; 162: 1367–1379.
Acknowledgements
Supported in part by the IZKF Jena, DFG (436 ARM 17/11/06, LI 820/15-1), DAAD (A0704616/Ref326), Ferring Pharmaceuticals and Organon France. We thank the Mapping Core and Map Finishing groups of the Wellcome Trust Sanger Institute for initial clone supply and verification.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bhatt, S., Moradkhani, K., Mrasek, K. et al. Breakpoint mapping and complete analysis of meiotic segregation patterns in three men heterozygous for paracentric inversions. Eur J Hum Genet 17, 44–50 (2009). https://doi.org/10.1038/ejhg.2008.144
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2008.144
Keywords
This article is cited by
-
Fitness consequences of polymorphic inversions in the zebra finch genome
Genome Biology (2016)
-
Molekularzytogenetische Methoden und Array-Diagnostik in der Pränatalmedizin
Medizinische Genetik (2011)