Abstract
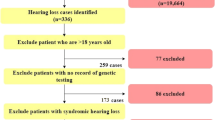
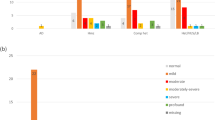
Hereditary hearing loss (HL) is a very heterogeneous trait, with 46 gene identifications for non-syndromic HL. Mutations in GJB2 cause up to half of all cases of severe-to-profound congenital autosomal recessive non-syndromic HL, with 35delG being the most frequent mutation in Caucasians. Although a genotype–phenotype correlation has been established for most GJB2 genotypes, the HL of 35delG homozygous patients is mild to profound. We hypothesise that this phenotypic variability is at least partly caused by the influence of modifier genes. By performing a whole-genome association (WGA) study on 35delG homozygotes, we sought to identify modifier genes. The association study was performed by comparing the genotypes of mild/moderate cases and profound cases. The first analysis included a pooling-based WGA study of a first set of 255 samples by using both the Illumina 550K and Affymetrix 500K chips. This analysis resulted in a ranking of all analysed single-nucleotide polymorphisms (SNPs) according to their P-values. The top 250 most significantly associated SNPs were genotyped individually in the same sample set. All 192 SNPs that still had significant P-values were genotyped in a second independent set of 297 samples for replication. The significant P-values were replicated in nine SNPs, with combined P-values between 3 × 10−3 and 1 × 10−4. This study suggests that the phenotypic variability in 35delG homozygous patients cannot be explained by the effect of one major modifier gene. Significantly associated SNPs may reflect a small modifying effect on the phenotype. Increasing the power of the study will be of greatest importance to confirm these results.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Morton CC, Nance WE : Newborn hearing screening – a silent revolution. N Engl J Med 2006; 354: 2151–2164.
Kenneson A, Van Naarden Braun K, Boyle C : GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: a HuGE review. Genet Med 2002; 4: 258–274.
Gasparini P, Rabionet R, Barbujani G et al: High carrier frequency of the 35delG deafness mutation in European populations. Genetic analysis consortium of GJB2 35delG. Eur J Hum Genet 2000; 8: 19–23.
Snoeckx RL, Huygen PL, Feldmann D et al: GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet 2005; 77: 945–957.
Bryda EC, Kim HJ, Legare ME, Frankel WN, Noben-Trauth K : High-resolution genetic and physical mapping of modifier-of-deafwaddler (mdfw) and Waltzer (Cdh23v). Genomics 2001; 73: 338–342.
Ikeda A, Zheng QY, Zuberi AR, Johnson KR, Naggert JK, Nishina PM : Microtubule-associated protein 1A is a modifier of tubby hearing (moth1). Nat Genet 2002; 30: 401–405.
Gabriel HD, Jung D, Butzler C et al: Transplacental uptake of glucose is decreased in embryonic lethal connexin26-deficient mice. J Cell Biol 1998; 140: 1453–1461.
Bykhovskaya Y, Mengesha E, Wang D et al: Phenotype of non-syndromic deafness associated with the mitochondrial A1555G mutation is modulated by mitochondrial RNA modifying enzymes MTO1 and GTPBP3. Mol Genet Metab 2004; 83: 199–206.
Craig DW, Huentelman MJ, Hu-Lince D et al: Identification of disease causing loci using an array-based genotyping approach on pooled DNA. BMC Genomics 2005; 6: 138.
Pearson JV, Huentelman MJ, Halperin RF et al: Identification of the genetic basis for complex disorders by use of pooling-based genomewide single-nucleotide-polymorphism association studies. Am J Hum Genet 2007; 80: 126–139.
Steer S, Abkevich V, Gutin A et al: Genomic DNA pooling for whole-genome association scans in complex disease: empirical demonstration of efficacy in rheumatoid arthritis. Genes Immun 2007; 8: 57–68.
Shifman S, Bhomra A, Smiley S et al: A whole genome association study of neuroticism using DNA pooling. Mol Psychiatry 2007; 13: 302–312.
Dunckley T, Huentelman MJ, Craig DW et al: Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med 2007; 357: 775–788.
Abecasis GR, Cherny SS, Cookson WO, Cardon LR : GRR: graphical representation of relationship errors. Bioinformatics 2001; 17: 742–743.
Curtis D, North BV, Gurling HM, Blaveri E, Sham PC : A quick and simple method for detecting subjects with abnormal genetic background in case–control samples. Ann Hum Genet 2002; 66: 235–244.
Lettre G, Lange C, Hirschhorn JN : Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol 2007; 31: 358–362.
Macgregor S, Zhao ZZ, Henders A, Nicholas MG, Montgomery GW, Visscher PM : Highly cost-efficient genome-wide association studies using DNA pools and dense SNP arrays. Nucleic Acids Res 2008; 36: e35.
Abe S, Koyama K, Usami S, Nakamura Y : Construction and characterization of a vestibular-specific cDNA library using T7-based RNA amplification. J Hum Genet 2003; 48: 142–149.
Nemoto T, Tanida I, Tanida-Miyake E et al: The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. J Biol Chem 2003; 278: 39517–39526.
Ohsumi Y : Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2: 211–216.
Libioulle C, Louis E, Hansoul S et al: Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet 2007; 3: e58.
Acknowledgements
We are very grateful to all patients who cooperated in this study. This study was supported in part by grants from EUROHEAR (LSHG-CT-2004-512063), the Fund for Scientific Research Flanders (FWO-F, Grant G.0138.07), the MNiSW Grant 0711/P01/2006/31, the Oticon Foundation (Denmark, MBP) and the National Institutes of Health (NIDCD DC02842 to RJHS). Nele Hilgert is a fellow of the Fund for Scientific Research Flanders (FWO-F).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hilgert, N., Huentelman, M., Thorburn, A. et al. Phenotypic variability of patients homozygous for the GJB2 mutation 35delG cannot be explained by the influence of one major modifier gene. Eur J Hum Genet 17, 517–524 (2009). https://doi.org/10.1038/ejhg.2008.201
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2008.201
Keywords
This article is cited by
-
Unusual phenotype in 35delG mutation: a case report
Journal of Medical Case Reports (2024)
-
Beyond Mendelian Inheritance: Genetic Buffering and Phenotype Variability
Phenomics (2022)
-
Characterization of GJB2 cis-regulatory elements in the DFNB1 locus
Human Genetics (2019)
-
Connexin 26 (GJB2) mutation in an Argentinean patient with keratitis-ichthyosis-deafness (KID) syndrome: a case report
BMC Medical Genetics (2016)
-
Mutations of GJB2 encoding connexin 26 contribute to non-syndromic moderate and severe hearing loss in Pakistan
European Archives of Oto-Rhino-Laryngology (2015)