Abstract
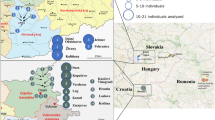
The human population is undergoing a major transition from a historical metapopulation structure of relatively isolated small communities to an outbred structure. This process is predicted to increase average individual genome-wide heterozygosity (h) and could have effects on health. We attempted to quantify this increase in mean h. We initially sampled 1001 examinees from a metapopulation of nine isolated villages on five Dalmatian islands (Croatia). Village populations had high levels of genetic differentiation, endogamy and consanguinity. We then selected 166 individuals with highly specific personal genetic histories to form six subsamples, which could be ranked a priori by their predicted level of outbreeding. The measure h was then estimated in the 166 examinees by genotyping 1184 STR/indel markers and using two different computation methods. Compared to the value of mean h in the least outbred sample, values of h in the remaining samples increased successively with predicted outbreeding by 0.023, 0.038, 0.058, 0.067 and 0.079 (P<0.0001), where these values are measured on the same scale as the inbreeding coefficient (but opposite sign). We have shown that urbanisation was associated with an average increase in h of up to 0.08–0.10 in this Croatian metapopulation, regardless of the method used. Similar levels of differentiation have been described in many populations. Therefore, changes in the level of heterozygosity across the genome of this magnitude may be common during isolate break-up in humans and could have significant health effects through the established genetic mechanism of hybrid vigour/heterosis.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Clark D : Urban World, Global City. UK: Routledge, 2004.
Darvasi A, Shifman S : The beauty of admixture. Nat Genet 2005; 37: 118–119.
Vitart V, Carothers AD, Suffolk R et al: Increased level of linkage disequilibrium in rural compared to urban communities: a factor to consider in association-study Design. Am J Hum Genet 2005; 76: 763–772.
Helgason A, Ingvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K : An Icelandic example of the impact of population structure on association studies. Nat Genet 2005; 37: 90–95.
Falconer DS, Mackay TFC : Introduction to Quantitative Genetics, 4th edn. Harlow, UK: Prentice Hall, 1996.
Charlesworth B, Hughes KA : Age-specific inbreeding depression and components of genetic variance in relation to the evolution of senescence. Proc Natl Acad Sci USA 1996; 93: 6140–6145.
Wright A, Charlesworth B, Rudan I, Carothers A, Campbell H : A polygenic basis for late-onset disease. Trends Genet 2003; 19: 97–106.
World Health Organisation: World Health Report 2002: Reducing Risks, Promoting Healthy Life. Geneva: WHO, 2002.
Abney M, McPeek MS, Ober C : Broad and narrow heritabilities of quantitative traits in a founder population. Am J Hum Genet 2001; 68: 1302–1307.
Rudan I, Campbell H, Carothers A et al: Inbreeding and the genetic complexity of human hypertension. Genetics 2003; 163: 1011–1021.
Rudan I, Campbell H, Carothers AD, Hastie ND, Wright AF : Contribution of consanguinity to polygenic and multifactorial diseases. Nat Genet 2006; 38: 1224–1225.
Rudan I, Campbell H : Five reasons why inbreeding may have considerable effect on post-reproductive human health. Coll Antropol 2004; 28: 943–950.
Bittles AH, Mason WM, Greene J, Rao NA : Reproductive behaviour and health in consanguineous marriages. Science 1991; 252: 789–794.
Bittles AH, Neel JV : The costs of human inbreeding and their implications for variations at the DNA level. Nat Genet 1994; 8: 117–121.
Vitart V, Biloglav Z, Hayward C et al: 3000 years of solitude: extreme level of differentiation in the island isolates of the Dalmatian coast. Eur J Hum Genet 2006; 14: 478–487.
Carothers AD, Rudan I, Hayward C et al: Estimating human individual inbreeding coefficients: comparison of genealogical and marker heterozygosity approaches. Ann Hum Genet 2006; 70: 666–676.
Campbell H, Carothers AD, Rudan I et al: Effects of genome-wide heterozygosity on a range of biomedically relevant human quantitative traits. Hum Mol Genet 2007; 16: 233–241.
Hoffman JI, Boyd IL, Amos W : Exploring the relationship between parental relatedness and male reproductive success in the Antarctic fur seal, Arctocephalus gazelle. Evolution 2004; 58: 2087–2099.
Coltman DW, Pilkington JG, Smith JA, Pemberton JM : Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution 1999; 53: 1259–1267.
Hirschhorn JN, Daly MJ : Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 2005; 6: 95–108.
Cavalli-Sforza LL, Feldman MW : Spatial subdivision of populations and estimates of genetic variation. Theor Popul Biol 1990; 37: 3–25.
Rosenberg NA, Pritchard JK, Weber JL et al: Genetic structure of human populations. Science 2002; 298: 2381–2385.
Charpentier M, Setchell JM, Prugnolle F : Genetic diversity and reproductive success in mandrills (Mandrillus sphinx). Proc Natl Acad Sci USA 2005; 102: 16723–16728.
Altukhov YP, Sheremet'eva VA, Rychkov YG : Heterosis as the cause of the secular trend in humans. Dokl Biol Sci 2000; 370: 43–46.
Mingroni MA : The secular rise in IQ: giving heterosis a closer look. Intelligence 2004; 32: 65–83.
Acknowledgements
Igor Rudan and Ozren Polasek were supported by Overseas Research Scheme and by the scholarship from the University of Edinburgh. Ozren Polasek is supported by the PhD scholarship from the University of Edinburgh. Igor Rudan and Zrinka Biloglav were supported by the British Scholarship Trust Fellowship. Zrinka Biloglav was supported by ‘Miroslav Cackovic’ fellowship of the Faculty of Medicine, University of Zagreb. The study was partially supported through the grants from the National Institutes of Health to Igor Rudan, Harry Campbell and James L Weber; Ministry of Science, Education and Sport of the Republic of Croatia to Igor Rudan (number 108-1080315-0302), and the grants from The British Council, The Wellcome Trust, The Royal Society and Medical Research Council to Harry Campbell and Igor Rudan and European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947). The authors collectively thank medical students of the Faculty of Medicine, University of Zagreb, Croatia; local general practitioners and nurses in study populations; the employees of several other Croatian institutions, including but not limited to the Institute for Anthropological Research in Zagreb, Croatia; the University of Rijeka and Split, Croatia; Croatian Institute of Public Health; Institutes of Public Health in Split and Dubrovnik, Croatia for their individual help in planning and carrying out the field work related to the project.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests statement
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Rudan, I., Carothers, A., Polasek, O. et al. Quantifying the increase in average human heterozygosity due to urbanisation. Eur J Hum Genet 16, 1097–1102 (2008). https://doi.org/10.1038/ejhg.2008.48
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2008.48
Keywords
This article is cited by
-
Genome-wide meta-analysis identifies novel loci associated with parathyroid hormone level
Molecular Medicine (2018)
-
Runs of homozygosity: windows into population history and trait architecture
Nature Reviews Genetics (2018)
-
Pairomics, the omics way to mate choice
Journal of Human Genetics (2013)
-
Does inbreeding affect N-glycosylation of human plasma proteins?
Molecular Genetics and Genomics (2011)
-
Genes predict village of origin in rural Europe
European Journal of Human Genetics (2010)