Abstract
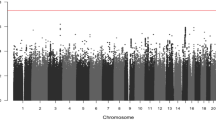
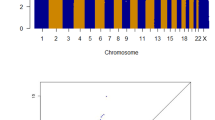
Genome scans based on gene-centric single nucleotide polymorphisms (SNPs) have been proposed as an efficient approach to identify disease-causing variants that is complementary to scans based on tagging SNPs. Adopting this approach to identify low-penetrance susceptibility alleles for colorectal cancer (CRC) we analysed genotype data from 9109 gene-centric SNPs, 7014 of which were non-synonymous (nsSNPs), in 2873 cases and 2871 controls using Illumina iselect arrays. Overall the distribution of associations was not significantly different from the null. No SNP achieved globally significant association after correction for multiple testing (lowest P value 1.7 × 10−4, rs727299). We then analysed the dataset incorporating information on the functional consequences of nsSNPs. We used results from the in silico algorithm PolyPhen as prior information to weight the association statistics, with weights estimated from the observed test statistics within predefined groups of SNPs. Incorporating this information did not, however, yield any further evidence of a specific association (lowest P value 2.2 × 10−4, rs1133950). There was a strong relationship between effect size and SNPs predicted to be damaging (P=1.63 × 10−5), however, these variants which are most likely to impact on risk are rare (MAF<5%). Hence although the rationale for searching for low-penetrance cancer susceptibly alleles by conducting genome-wide scans of coding changes is strong, in practice it is likely that natural selection has rendered such alleles to be too rare to be detected by association studies of the size employed.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Lichtenstein P, Holm NV, Verkasalo PK et al: Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 2000; 343: 78–85.
Aaltonen L, Johns L, Jarvinen H, Mecklin JP, Houlston R : Explaining the familial colorectal cancer risk associated with mismatch repair (MMR)-deficient and MMR-stable tumors. Clin Cancer Res 2007; 13: 356–361.
Zanke BW, Greenwood CM, Rangrej J et al: Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat Genet 2007; 39: 989–994.
Tenesa A, Farrington SM, Prendergast JG et al: Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat Genet 2008; 40: 631–637.
Tomlinson I, Webb E, Carvajal-Carmona L et al: A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat Genet 2007; 39: 984–988.
Tomlinson IP, Webb E, Carvajal-Carmona L et al: A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet 2008; 40: 623–630.
Jaeger E, Webb E, Howarth K et al: Common genetic variants at the CRAC1 (HMPS) locus on chromosome 15q13.3 influence colorectal cancer risk. Nat Genet 2008; 40: 26–28.
Broderick P, Carvajal-Carmona L, Pittman AM et al: A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet 2007; 39: 1315–1317.
Botstein D, Risch N : Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 2003; 33 (Suppl): 228–237.
Burton PR, Clayton DG, Cardon LR et al: Association scan of 14 500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet 2007; 39: 1329–1337.
Penegar S, Wood W, Lubbe S et al: National study of colorectal cancer genetics. Br J Cancer 2007; 97: 1305–1309.
Eisen T, Matakidou A, Consortium G, Houlston R : Identification of low penetrance alleles for lung cancer: the GEnetic Lung CAncer Predisposition Study (GELCAPS). BMC Cancer 2008; 8: 244.
Ramensky V, Bork P, Sunyaev S : Human non-synonymous SNPs: server and survey. Nucleic Acids Res 2002; 30: 3894–3900.
Xi T, Jones IM, Mohrenweiser HW : Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics 2004; 83: 970–979.
Roeder K, Devlin B, Wasserman L : Improving power in genome-wide association studies: weights tip the scale. Genet Epidemiol 2007; 31: 741–747.
Hochberg Y, Benjamini Y : More powerful procedures for multiple significance testing. Stat Med 1990; 9: 811–818.
Hague A, Hicks DJ, Hasan F et al: Increased sensitivity to TRAIL-induced apoptosis occurs during the adenoma to carcinoma transition of colorectal carcinogenesis. Br J Cancer 2005; 92: 736–742.
Cybulski C, Gorski B, Huzarski T et al: CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 2004; 75: 1131–1135.
Cox A, Dunning AM, Garcia-Closas M et al: A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet 2007; 39: 352–358.
Cargill M, Altshuler D, Ireland J et al: Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet 1999; 22: 231–238.
Leabman MK, Huang CC, DeYoung J et al: Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci USA 2003; 100: 5896–5901.
Swift M, Morrell D, Massey RB, Chase CL : Incidence of cancer in 161 families affected by ataxia-telangiectasia. N Engl J Med 1991; 325: 1831–1836.
Renwick A, Thompson D, Seal S et al: ATM mutations that cause ataxia-telangiectasia are breast cancer susceptibility alleles. Nat Genet 2006; 38: 873–875.
Seal S, Thompson D, Renwick A et al: Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet 2006; 38: 1239–1241.
Acknowledgements
Bobby Moore Cancer Research UK provided principal funding for this study. Additional funding was provided by the European Union (CPRB LSHC-CT-2004-503465) and CORE. IC was in receipt of a clinical training fellowship from St George's Hospital Medical School. We are grateful to colleagues within the UK National Cancer Research Network and to Stephen Penegar for his diligence in supporting the NSCCG. Finally, we thank all individuals who participated in this study.
Author information
Authors and Affiliations
Corresponding author
Additional information
Conflict of interest
The authors declare no conflict of interest.
URLs
Illumina: http://www.illumina.com/
Online Inheritance in Man: http://www.ncbi.nlm.nih.gov/sites/entrez
The R suite can be found at http://www.r-project.org/
dbSNP: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?CMD=search&DB=snp
GELCAPS: http://pfsearch.ukcrn.org.uk/StudyDetail.aspx?TopicID=1&StudyID=781
-http://www.dh.gov.uk/assetRoot/04/01/45/13/04014513.pdf
National Study of Colorectal Cancer Genetics (NSCCG): http://pfsearch.ukcrn.org.uk/StudyDetail.aspx?TopicID=1&StudyID=1269
Polyphen: http://genetics.bwh.harvard.edu/pph/data/index.html
Rights and permissions
About this article
Cite this article
Webb, E., Broderick, P., Lubbe, S. et al. A genome-wide scan of 10 000 gene-centric variants and colorectal cancer risk. Eur J Hum Genet 17, 1507–1514 (2009). https://doi.org/10.1038/ejhg.2009.92
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2009.92
Keywords
This article is cited by
-
Genome-wide scan of the effect of common nsSNPs on colorectal cancer survival outcome
British Journal of Cancer (2018)
-
Association genetics of phenolic needle compounds in Norway spruce with variable susceptibility to needle bladder rust
Plant Molecular Biology (2017)
-
Association study identifying polymorphisms in CD47 and other extracellular matrix pathway genes as putative prognostic markers for colorectal cancer
International Journal of Colorectal Disease (2013)