Abstract
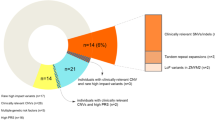
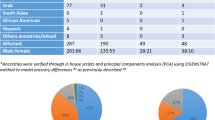
Recent reports suggest that somatic structural changes occur in the human genome, but how these genomic alterations might contribute to disease is unknown. Using samples collected as part of the International Schizophrenia Consortium (schizophrenia, n=3518; control, n=4238) recruited across multiple university research centers, we assessed single-nucleotide polymorphism genotyping arrays for evidence of chromosomal anomalies. Data from genotyping arrays on each individual were processed using Birdsuite and analyzed with PLINK. We validated potential chromosomal anomalies using custom nanostring probes and quantitative PCR. We estimate chromosomal alterations in the schizophrenia population to be 0.42%, which is not significantly different from controls (0.26%). We identified and validated a set of four extremely large (>10 Mb) chromosomal anomalies in subjects with schizophrenia, including a chromosome 8 trisomy and deletion of the q arm of chromosome 7. These data demonstrate that chromosomal anomalies are present at low frequency in blood cells of both control and schizophrenia subjects.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Kendler KS, Gallagher TJ, Abelson JM, Kessler RC : Lifetime prevalence, demographic risk factors, and diagnostic validity of nonaffective psychosis as assessed in a US community sample. The National Comorbidity Survey. Arch Gen Psychiatry 1996; 53: 1022–1031.
Cannon TD, Kaprio J, Lonnqvist J, Huttunen M, Koskenvuo M : The genetic epidemiology of schizophrenia in a Finnish twin cohort. A population-based modeling study. Arch Gen Psychiatry 1998; 55: 67–74.
International Schizophrenia Consortium: Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 2008; 455: 237–241.
Purcell SM, Wray NR, Stone JL et al: Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
Shi J, Levinson DF, Duan J et al: Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009; 460: 753–757.
Mulle JG, Dodd AF, McGrath JA et al: Microdeletions of 3q29 confer high risk for schizophrenia. Am J Hum Genet 2010; 87: 229–236.
Vacic V, McCarthy S, Malhotra D et al: Duplications of the neuropeptide receptor gene VIPR2 confer significant risk for schizophrenia. Nature 2011; 471: 499–503.
Brunet A, Armengol L, Heine D et al: BAC array CGH in patients with velocardiofacial syndrome-like features reveals genomic aberrations on chromosome region 1q21.1. BMC Med Genet 2009; 10: 144.
Bassett AS, Scherer SW, Brzustowicz LM : Copy number variations in schizophrenia: critical review and new perspectives on concepts of genetics and disease. Am J Psychiatry 2010; 167: 899–914.
Forsberg LA, Rasi C, Razzaghian HR et al: Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet 2012; 90: 217–228.
Rodriguez-Santiago B, Malats N, Rothman N et al: Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am J Hum Genet 2010; 87: 129–138.
Jacobs KB, Yeager M, Zhou W et al: Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012; 44: 651–658.
Bergen SE, O’Dushlaine CT, Ripke S et al: Genome-wide association study in a Swedish population yields support for greater CNV and MHC involvement in schizophrenia compared with bipolar disorder. Mol Psychiatry 2012; 17: 880–886.
Korn JM, Kuruvilla FG, McCarroll SA et al: Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet 2008; 40: 1253–1260.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Ye T, Lipska BK, Tao R et al: Analysis of copy number variations in brain DNA from patients with schizophrenia and other psychiatric disorders. Biol Psychiatry 2012; 72: 651–654.
Wilson GM, Flibotte S, Chopra V, Melnyk BL, Honer WG, Holt RA : DNA copy-number analysis in bipolar disorder and schizophrenia reveals aberrations in genes involved in glutamate signaling. Hum Mol Genet 2006; 15: 743–749.
Acknowledgements
We thank all subjects for contributing DNA to our research centers.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Ruderfer, D., Chambert, K., Moran, J. et al. Mosaic copy number variation in schizophrenia. Eur J Hum Genet 21, 1007–1011 (2013). https://doi.org/10.1038/ejhg.2012.287
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2012.287
Keywords
This article is cited by
-
ARFID Genes and Environment (ARFID-GEN): study protocol
BMC Psychiatry (2023)
-
Genome-wide association study of copy number variation with lung function identifies a novel signal of association near BANP for forced vital capacity
BMC Genetics (2016)
-
Copy number variation in schizophrenia in Sweden
Molecular Psychiatry (2014)