Abstract
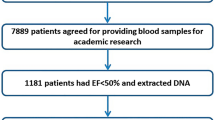
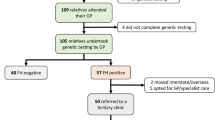
In recent years, multiple loci dispersed on the genome have been shown to be associated with coronary artery disease (CAD). We investigated whether these common genetic variants also hold value for CAD prediction in a large cohort of patients with familial hypercholesterolemia (FH). We genotyped a total of 41 single-nucleotide polymorphisms (SNPs) in 1701 FH patients, of whom 482 patients (28.3%) had at least one coronary event during an average follow up of 66 years. The association of each SNP with event-free survival time was calculated with a Cox proportional hazard model. In the cardiovascular disease risk factor adjusted analysis, the most significant SNP was rs1122608:G>T in the SMARCA4 gene near the LDL-receptor (LDLR) gene, with a hazard ratio for CAD risk of 0.74 (95% CI 0.49–0.99; P-value 0.021). However, none of the SNPs reached the Bonferroni threshold. Of all the known CAD loci analyzed, the SMARCA4 locus near the LDLR had the strongest negative association with CAD in this high-risk FH cohort. The effect is contrary to what was expected. None of the other loci showed association with CAD.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Goldstein JL, Sobhani MK, Faust JR et al: Heterozygous familial hypercholesterolemia: failure of normal allele to compensate for mutant allele at a regulated genetic locus. Cell 1976; 9: 195–203.
Huijgen R, Vissers MN, Defesche JC et al: Familial hypercholesterolemia: current treatment and advances in management. Expert Rev Cardiovasc Ther 2008; 6: 567–581.
Van Aalst-Cohen ES, Jansen AC, Tanck MW et al: Diagnosing familial hypercholesterolaemia: the relevance of genetic testing. Eur Heart J 2006; 27: 2240–2246.
Stone NJ, Levy RI, Fredrickson DS et al: Coronary artery disease in 116 kindred with familial type II hyperlipoproteinemia. Circulation 1974; 49: 476–488.
Sijbrands EJ, Westendorp RG, Defesche JC et al: Mortality over two centuries in large pedigree with familial hypercholesterolaemia: family tree mortality study. BMJ 2001; 322: 1019–1023.
Jansen AC, van Aalst-Cohen ES, Tanck MW et al: The contribution of classical risk factors to cardiovascular disease in familial hypercholesterolaemia: data in 2400 patients. J Intern Med 2004; 256: 482–490.
Souverein OW, Defesche JC, Zwinderman AH et al: Influence of LDL-receptor mutation type on age at first cardiovascular event in patients with familial hypercholesterolaemia. Eur Heart J 2007; 28: 299–304.
Jansen AC, van Aalst-Cohen ES, Tanck MW et al: The contribution of classical risk factors to cardiovascular disease in familial hypercholesterolaemia: data in 2400 patients. J Intern Med 2004; 256: 482–490.
Peden JF, Hopewell JC, Saleheen D et al: A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat Genet 2011; 43: 339–344.
Schunkert H, Konig IR, Kathiresan S et al: Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 2011; 43: 333–338.
Sivapalaratnam S, Motazacker MM, Maiwald S et al: Genome-wide association studies in atherosclerosis. Curr Atheroscler Rep 2011; 13: 225–232.
Deloukas P, Kanoni S, Willenborg C et al: Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2012; 45: 25–33.
Umans-Eckenhausen MA, Defesche JC, Sijbrands EJ et al: Review of first 5 years of screening for familial hypercholesterolaemia in the Netherlands. Lancet 2001; 357: 165–168.
Jansen AC, van Aalst-Cohen ES, Tanck MW et al: Genetic determinants of cardiovascular disease risk in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol 2005; 25: 1475–1481.
Hao K, Chudin E, McElwee J et al: Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet 2009; 10: 27.
Li Y, Willer CJ, Ding J et al: MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol 2010; 34: 816–834.
Southam L, Panoutsopoulou K, Rayner NW et al: The effect of genome-wide association scan quality control on imputation outcome for common variants. Eur J Hum Genet 2011; 19: 610–614.
Keating BJ, Tischfield S, Murray SS et al: Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS One 2008; 3: e3583.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Aulchenko YS, Struchalin MV, van Duijn CM : ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics 2010; 11: 134.
Jansen AC, van Aalst-Cohen ES, Tanck MW et al: Genetic determinants of cardiovascular disease risk in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol 2005; 25: 1475–1481.
Khot UN, Khot MB, Bajzer CT et al: Prevalence of conventional risk factors in patients with coronary heart disease. JAMA 2003; 290: 898–904.
Muehlschlegel JD, Liu KY, Perry TE et al: Chromosome 9p21 variant predicts mortality after coronary artery bypass graft surgery. Circulation 2010; 122: S60–S65.
Gong Y, Beitelshees AL, Cooper-Dehoff RM et al: Chromosome 9p21 haplotypes and prognosis in white and black patients with coronary artery disease. Circ Cardiovasc Genet 2011; 4: 169–178.
Martinelli N, Girelli D, Lunghi B et al: Polymorphisms at LDLR locus may be associated with coronary artery disease through modulation of coagulation factor VIII activity and independently from lipid profile. Blood 2010; 116: 5688–5697.
Murabito JM, White CC, Kavousi M et al: Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ Cardiovasc Genet 2012; 5: 100–112.
Kathiresan S, Voight BF, Purcell S et al: Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009; 41: 334–341.
Ripatti S, Tikkanen E, Orho-Melander M et al: A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet 2010; 376: 1393–1400.
Paynter NP, Chasman DI, Buring JE et al: Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med 2009; 150: 65–72.
Acknowledgements
This work was supported by Bloodomics consortium, European Union 6th Framework Programme (LSHM-CT-2004-503485), Ipse Movet, the Lifetime Achievement Award of the Dutch Heart Foundation (2010T082) to JJPK and grants from the British Heart Foundation and the National Institute for Health Research, England to WHO. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
van Iperen, E., Sivapalaratnam, S., Boekholdt, S. et al. Common genetic variants do not associate with CAD in familial hypercholesterolemia. Eur J Hum Genet 22, 809–813 (2014). https://doi.org/10.1038/ejhg.2013.242
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.242
Keywords
This article is cited by
-
Progress in the care of common inherited atherogenic disorders of apolipoprotein B metabolism
Nature Reviews Endocrinology (2016)