Abstract
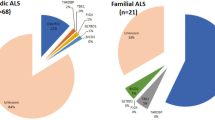
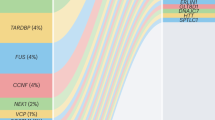
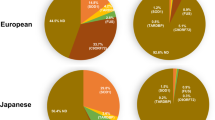
Amyotrophic lateral sclerosis (ALS) may appear to be familial or sporadic, with recognised dominant and recessive inheritance in a proportion of cases. Sporadic ALS may be caused by rare homozygous recessive mutations. We studied patients and controls from the UK and a multinational pooled analysis of GWAS data on homozygosity in ALS to determine any potential recessive variant leading to the disease. Six-hundred and twenty ALS and 5169 controls were studied in the UK cohort. A total of 7646 homozygosity segments with length >2 Mb were identified, and 3568 rare segments remained after filtering ‘common’ segments. The mean total of the autosomal genome with homozygosity segments was longer in ALS than in controls (unfiltered segments, P=0.05). Two-thousand and seventeen ALS and 6918 controls were studied in the pooled analysis. There were more regions of homozygosity segments per case (P=1 × 10−5), a greater proportion of cases harboured homozygosity (P=2 × 10−5), a longer average length of segment (P=1 × 10−5), a longer total genome coverage (P=1 × 10−5), and a higher rate of these segments overlapped with RefSeq gene regions (P=1 × 10−5), in ALS patients than controls. Positive associations were found in three regions. The most significant was in the chromosome 21 SOD1 region, and also chromosome 1 2.9–4.8 Mb, and chromosome 5 in the 65 Mb region. There are more than twenty potential genes in these regions. These findings point to further possible rare recessive genetic causes of ALS, which are not identified as common variants in GWAS.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Byrne S, Walsh C, Lynch C et al: Rate of familial amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry 2011; 82: 623–627.
Rosen DR, Siddique T, Patterson D et al: Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993; 362: 59–62, (Erratum in: Nature; 364: 362).
Sreedharan J, Blair IP, Tripathi VB et al: TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008; 319: 1668–1672.
Vance C, Rogelj B, Hortobágyi T et al: Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009; 323: 1208–1211.
Johnson JO, Mandrioli J, Benatar M et al: Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010; 68: 857–864, (Erratum in: Neuron 2011;69:397).
Renton AE, Majounie E, Waite A et al: A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011; 72: 257–268.
DeJesus-Hernandez M, Mackenzie IR, Boeve BF et al: Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011; 72: 245–256.
Andersen PM, Nilsson P, Ala-Hurula V et al: Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat Genet 1995; 10: 61–66.
Yang Y, Hentati A, Deng HX et al: The gene encoding alsin, a protein with three guanine-nucleotide exchange factor domains, is mutated in a form of recessive amyotrophic lateral sclerosis. Nat Genet 2001; 29: 160–165.
Hadano S, Hand CK, Osuga H et al: A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 2001; 29: 166–173.
Maruyama H, Morino H, Ito H et al: Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010; 465: 223–226.
Nalls MA, Simon-Sanchez J, Gibbs JR et al: Measures of autozygosity in decline: globalization, urbanization, and its implications for medical genetics. PLoS Genet 2009; 5: e1000415.
Brooks BR, Miller RG, Swash M, Munsat TL : World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord 2000; 1: 293–299.
Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven com mon diseases and 3,000 shared controls. Nature 2007; 447: 661–678.
Shatunov A, Mok K, Newhouse S et al: Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol 2010; 9: 986–994, (Erratum in: Lancet Neurol 2011; 10: 205).
Laaksovirta H, Peuralinna T, Schymick JC et al: Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol 2010; 9: 978–985.
Cronin S, Berger S, Ding J et al: A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet 2008; 17: 768–774.
Schymick JC, Scholz SW, Fung H et al: Genome-wide genotyping in amyotrophic lateral sclerosis and neurologically normal controls: first stage analysis and public release of data. Lancet Neurol 2007; 6: 322–328.
Chiò A, Schymick JC, Restagno G et al: A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum Mol Genet 2009; 18: 1524–1532.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
HapMap, ‘HapMapIII Release 2010-05’, February 2010, http://hapmap.ncbi.nlm.nih.gov/downloads/genotypes/2010-05_phaseIII/plink_format/..
Illumina, ‘Genome-Wide DNA Analysis Beadchips’ 2010, http://www.illumina.com/Documents/products/datasheets/datasheet_infiniumhd.pdf. http://www.illumina.com/Documents/products/datasheets/datasheet_omni_whole-genome_arrays.pdf. http://www.illumina.com/Documents/products/technotes/technote_intelligent_snp_selection.pdf. http://www.solexa.co.uk/downloads/HUMANHAP550_Duo_DataSheet.pdf.
Sabatti C, Risch N : Homozygosity and linkage disequilibrium. Genetics 2002; 160: 1707–1719.
Wang H, Lin CH, Service S, Chen Y, Freimer N, Sabatti C : Linkage disequilibrium and haplotype homozygosity in population samples genotyped at a high marker density. Hum Hered 2006; 62: 175–189.
R Development Core Team. 2008: R: a language and environment for statistical computing, in: R Development Core Team (ed.) Vienna, Austria: R Foundation for Statistical Computing, 2008.
‘NCBI Gene’ 2011, http://www.ncbi.nlm.nih.gov/gene..
Andersen PM, Al-Chalabi A : Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol 2011; 11: 603–615.
Lai C, Xie C, McCormack SG et al: Amyotrophic lateral sclerosis 2-deficiency leads to neuronal degeneration in amyotrophic lateral sclerosis through altered AMPA receptor trafficking. J Neurosci 2006; 26: 11798–11806.
Hadano S, Otomo A, Kunita R et al: Loss of ALS2/Alsin exacerbates motor dysfunction in a SOD1-expressing mouse ALS model by disturbing endolysosomal trafficking. PLoS ONE 2010; 5: e9805.
Li Q, Spencer NY, Pantazis NJ, JF Engelhardt : Alsin and SOD1(G93A) proteins regulate endosomal reactive oxygen species production by glial cells and proinflammatory pathways responsible for neurotoxicity. J Biol Chem 2011; 286: 40151–40162.
Osawa T, Mizuno Y, Fujita Y, Takatama M, Nakazato Y, Okamoto K : Optineurin in neurodegenerative diseases. Neuropathology 2011; 31: 569–574.
Ito H, Fujita K, Nakamura M et al: Optineurin is co-localized with FUS in basophilic inclusions of ALS with FUS mutation and in basophilic inclusion body disease. Acta Neuropathol 2011; 121: 555–557.
Sugihara K, Maruyama H, Kamada M, Morino H, Kawakami H : Screening for OPTN mutations in amyotrophic lateral sclerosis in a mainly Caucasian population. Neurobiol Aging 2011; 32: 1923 e9–10.
Al-Chalabi A, Andersen PM, Chioza et al: Recessive amyotrophic lateral sclerosis families with the D90A SOD1 mutation share a common founder: evidence for a linked protective factor. Hum Mol Genet 1998; 7: 2045–2050.
Parton MJ, Broom W, Andersen PM et al: D90A-SOD1 mediated amyotrophic lateral sclerosis: a single founder for all cases with evidence for a Cis-acting disease modifier in the recessive haplotype. Hum Mutat 2002; 20: 473.
Casey JP, Magalhaes T, Conroy JM et al: A novel approach of homozygous haplotype sharing identifies candidate genes in autism spectrum disorder. Hum Genet 2012; 131: 565–579.
Cox LE, Ferraiuolo L, Goodall EF et al: Mutations in CHMP2B in lower motor neuron predominant amyotrophic lateral sclerosis (ALS). PLoS ONE 2010; 5: e9872.
Sexton T, Bantignies F, Cavalli G : Genomic interactions: chromatin loops and gene meeting points in transcriptional regulation. Semin Cell Dev Biol 2009; 20: 849–855.
Wellcome Trust Sanger Institute, ‘snoU13’ 2011, http://rfam.sanger.ac.uk/family/RF01210..
Acknowledgements
This work was supported in part by the Intramural Research Programs of the NIH, the National Institute on Aging (Z01-AG000949-02), and the National Institute of Neurological Disorders and Stroke. Extramural NIH grants R01AG031278 and R01AG038791 supported some family assessments. The research leading to these results has received funding from the European Community’s Health Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 259867. We thank the Motor Neurone Disease Association of Great Britain for several grants relating to this work (RWO, AAC, PJS, HM), the ALS Association, The Angel Fund, the ALS Therapy Alliance, and the Wellcome Trust (PJS) for support. This work was also funded by the Reta Lila Weston Foundation, and by an MRC returning scientist (JH) and fellowship (SPB) award, by Microsoft Research Foundation, the ALS Association, Helsinki University Central Hospital, the Finnish Academy, Ministero della Salute, Progetti Finalizzati 2007, Fondazione Vialli e Mauro for ALS, and Federazione Italiana Giuoco Calcio. The authors thank the NIHR specialist Biomedical Research Centre for Mental Health at the South London and Maudsley NHS Foundation Trust (SLaM) and the Institute of Psychiatry, King’s College London, and the National Institute for Health Research-funded University College London / University College London Hospitals Biomedical Research Centre.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on European Journal of Human Genetics website
Supplementary information
Rights and permissions
About this article
Cite this article
Mok, K., Laaksovirta, H., Tienari, P. et al. Homozygosity analysis in amyotrophic lateral sclerosis. Eur J Hum Genet 21, 1429–1435 (2013). https://doi.org/10.1038/ejhg.2013.59
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2013.59
Keywords
This article is cited by
-
CRISPR-mediated gene correction links the ATP7A M1311V mutations with amyotrophic lateral sclerosis pathogenesis in one individual
Communications Biology (2020)
-
Identification of a de novo DYNC1H1 mutation via WES according to published guidelines
Scientific Reports (2016)
-
Establishing the UK DNA Bank for motor neuron disease (MND)
BMC Genetics (2015)
-
Inbreeding and homozygosity in breast cancer survival
Scientific Reports (2015)
-
Exome sequencing of case-unaffected-parents trios reveals recessive and de novo genetic variants in sporadic ALS
Scientific Reports (2015)