Abstract
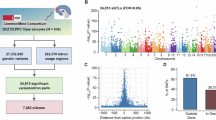
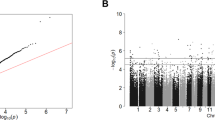
Emerging evidence suggests that schizophrenia (SZ) susceptibility involves variation at genetic, epigenetic and transcriptome levels. We describe an integrated approach that leverages DNA methylation and gene expression data to prioritize genetic variation involved in disease. DNA methylation levels were obtained from whole blood of 260 SZ patients and 250 unaffected controls of which a subset with gene expression data was available. By assessing DNA methylation and gene expression in cases and controls, we identified 432 CpG sites with differential methylation levels that are associated with differential gene expression. We hypothesized that genetic factors involved in these methylation levels may be associated with the genetic risk of SZ susceptibility. To test this hypothesis, we used results from the Psychiatric Genomics Consortium SZ genome-wide association study (GWAS). We observe an enrichment of SZ-associated SNPs in the mQTLs of which the associated CpG site is also correlated with differential gene expression in SZ. While this enrichment was already apparent when using nominal significant thresholds, enrichment was even more pronounced when applying more stringent significance levels. One locus, previously identified as susceptibility locus in a SZ GWAS, involves SNP rs11191514:C>T, which regulates DNA methylation of calcium homeostasis modulator 1 that is also associated with differential gene expression in patients. Overall, our results suggest that epigenetic variation plays an important role in SZ susceptibility and that the integration of analyses of genetic, epigenetic and gene expression profiles may be a biologically meaningful approach for identifying disease susceptibility loci, even when using whole blood data in studies of brain-related disorders.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Lee SH, DeCandia TR, Ripke S et al: Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet 2012; 44: 247–250.
Sullivan PF, Daly MJ, O'Donovan M : Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet 2012; 13: 537–551.
Sullivan PF, Kendler KS, Neale MC : Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry 2003; 60: 1187–1192.
Purcell SM, Wray NR, Stone JL et al: Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009; 460: 748–752.
So HC, Gui AH, Cherny SS, Sham PC : Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol 2011; 35: 310–317.
Manolio TA, Collins FS, Cox NJ et al: Finding the missing heritability of complex diseases. Nature 2009; 461: 747–753.
Petronis A : Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature 2010; 465: 721–727.
Labrie V, Pai S, Petronis A : Epigenetics of major psychosis: progress, problems and perspectives. Trends Genet 2012; 28: 427–435.
Feinberg AP, Irizarry RA : Evolution in health and medicine Sackler colloquium: Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci USA 2010; 107 (Suppl 1): 1757–1764.
Feinberg AP : Phenotypic plasticity and the epigenetics of human disease. Nature 2007; 447: 433–440.
Bjornsson HT, Fallin MD, Feinberg AP : An integrated epigenetic and genetic approach to common human disease. Trends Genet 2004; 20: 350–358.
Richards AL, Jones L, Moskvina V et al: Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry 2012; 17: 193–201.
Gamazon ER, Badner JA, Cheng L et al: Enrichment of cis-regulatory gene expression SNPs and methylation quantitative trait loci among bipolar disorder susceptibility variants. Mol Psychiatry 2012; 18: 340–346.
Ripke S, Sanders AR, Kendler KS et al: Genome-wide association study identifies five new schizophrenia loci. Nat Genet 2011; 43: 969–976.
Smyth GK : Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004; 3: Article3.
Horvath S, Zhang Y, Langfelder P et al: Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol 2012; 13: R97.
van Eijk KR, de Jong S, Boks MP et al: Genetic analysis of DNA methylation and gene expression levels in whole blood of healthy human subjects. BMC Genomics 2012; 13: 636.
de Jong S, van Eijk KR, Zeegers DW et al: Expression QTL analysis of top loci from GWAS meta-analysis highlights additional schizophrenia candidate genes. Eur J Hum Genet 2012; 20: 1004–1008.
Purcell S, Neale B, Todd-Brown K et al: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Dreses-Werringloer U, Lambert JC, Vingtdeux V et al: A polymorphism in CALHM1 influences Ca2+ homeostasis, Abeta levels, and Alzheimer's disease risk. Cell 2008; 133: 1149–1161.
Harrison PJ : The hippocampus in schizophrenia: a review of the neuropathological evidence and its pathophysiological implications. Psychopharmacology 2004; 174: 151–162.
Ripke S, O'Dushlaine C, Chambert K et al: Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet 2013; 45: 1150–1159.
Stefansson H, Ophoff RA, Steinberg S et al: Common variants conferring risk of schizophrenia. Nature 2009; 460: 744–747.
Yoo YA, Kim MJ, Park JK et al: Mitochondrial ribosomal protein L41 suppresses cell growth in association with p53 and p27Kip1. Mol Cell Biol 2005; 25: 6603–6616.
Acknowledgements
We thank the patients and controls for participating in the study. This study was supported by National Institutes of Health (NIH) grants R01 DA028526, R01 NS058980 and RO1 MH 078075 (to RAO). We thank Dr Kim Staats for critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
van Eijk, K., de Jong, S., Strengman, E. et al. Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood. Eur J Hum Genet 23, 1106–1110 (2015). https://doi.org/10.1038/ejhg.2014.245
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2014.245
This article is cited by
-
Meta-analysis of epigenetic aging in schizophrenia reveals multifaceted relationships with age, sex, illness duration, and polygenic risk
Clinical Epigenetics (2024)
-
A meta-analysis of immune-cell fractions at high resolution reveals novel associations with common phenotypes and health outcomes
Genome Medicine (2023)
-
A multimodal attempt to follow-up linkage regions using RNA expression, SNPs and CpG methylation in schizophrenia and bipolar disorder kindreds
European Journal of Human Genetics (2020)
-
Epigenetic mechanisms in schizophrenia and other psychotic disorders: a systematic review of empirical human findings
Molecular Psychiatry (2020)
-
Longitudinal investigation of DNA methylation changes preceding adolescent psychotic experiences
Translational Psychiatry (2019)