Abstract
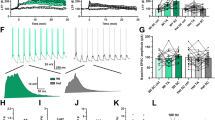
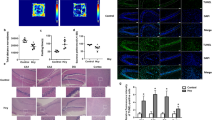
Learning disabilities (LDs) are a clinically and genetically heterogeneous group of diseases. Array-CGH and high-throughput sequencing have dramatically expanded the number of genes implicated in isolated intellectual disabilities and LDs, highlighting the implication of neuron-specific post-mitotic transcription factors and synaptic proteins as candidate genes. We report a unique family diagnosed with autosomal dominant learning disability and a 6p21 microdeletion segregating in three patients. The 870 kb microdeletion encompassed the brain-expressed gene LRFN2, which encodes for a synaptic cell adhesion molecule. Neuropsychological assessment identified selective working memory deficits, with borderline intellectual functioning. Further investigations identified a defect in executive function, and auditory-verbal processes. These data were consistent with brain MRI and FDG-PET functional brain imaging, which, when compared with controls, revealed abnormal brain volume and hypometabolism of gray matter structures implicated in working memory. We performed electron microscopy immunogold labeling demonstrating the localization of LRFN2 at synapses of cerebellar and hippocampal rat neurons, often associated with the NR1 subunit of N-methyl-D-aspartate receptors (NMDARs). Altogether, the combined approaches imply a role for LRFN2 in LD, specifically for working memory processes and executive function. In conclusion, the identification of familial cases of clinically homogeneous endophenotypes of LD might help in both the management of patients and genetic counseling for families.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Flint J : The genetic basis of cognition. Brain J Neurol 1999; 122: 2015–2032.
Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP : A forkhead-domain gene is mutated in a severe speech and language disorder. Nature 2001; 413: 519–523.
Graham SA, Fisher SE : Decoding the genetics of speech and language. Curr Opin Neurobiol 2013; 23: 43–51.
Newbury DF, Fisher SE, Monaco AP : Recent advances in the genetics of language impairment. Genome Med 2010; 2: 6.
Baddeley AD, Hitch GJ : Development of working memory: should the Pascual-Leone and the Baddeley and Hitch models be merged? J Exp Child Psychol 2000; 77: 128–137.
Baddeley A : The concept of working memory: a view of its current state and probable future development. Cognition 1981; 10: 17–23.
Khan ZU, Muly EC : Molecular mechanisms of working memory. Behav Brain Res 2011; 219: 329–341.
Holmes J, Gathercole SE, Dunning DL : Poor working memory: impact and interventions. Adv Child Dev Behav 2010; 39: 1–43.
Gathercole SE, Alloway TP : Practitioner review: short-term and working memory impairments in neurodevelopmental disorders: diagnosis and remedial support. J Child Psychol Psychiatry 2006; 47: 4–15.
Gathercole SE, Alloway TP, Willis C, Adams A-M : Working memory in children with reading disabilities. J Exp Child Psychol 2006; 93: 265–281.
Kaufman AS, Lichtenberger EO : Assessing Adolescent and Adult Intelligence. John Wiley & Sons, 2005.
Bennett SJ, Holmes J, Buckley S : Computerized memory training leads to sustained improvement in visuospatial short-term memory skills in children with Down syndrome. Am J Intellect Dev Disabil 2013; 118: 179–192.
Wechsler D : Wechsler memory scale (WMS-III). Psychological Corporation, 1997.
Zimmermann P, Fimm B : Test for attentional performance (TAP). PsyTest Herzogenrath, 1995.
Murphy MD, Schmitt FA, Caruso MJ, Sanders RE : Metamemory in older adults: the role of monitoring in serial recall. Psychol Aging 1987; 2: 331–339.
Case R, Kurland DM, Goldberg J : Operational efficiency and the growth of short-term memory span. J Exp Child Psychol 1982; 33: 386–404.
Conway AR, Cowan N, Bunting MF, Therriault DJ, Minkoff SR : A latent variable analysis of working memory capacity, short-term memory capacity, processing speed, and general fluid intelligence. Intelligence 2002; 30: 163–183.
Brickenkamp R : Test d2. Concentration-Endurence Test: Manual, 5th edn. Gottingen, Germany: Verlag für Psychologie, 1981.
Reitan RM : Trail Making Test: Manual for administration and scoring. Reitan Neuropsychology Laboratory, 1986.
Heaton RK : A manual for the Wisconsin card sorting test. Western Psycological Services, 1981.
Petralia RS, Wenthold RJ : Immunocytochemistry of NMDA receptors. Methods Mol Biol 1999; 128: 73–92.
Petralia RS : Distribution of extrasynaptic NMDA receptors on neurons. ScientificWorldJournal 2012; 2012: 267120.
Petralia RS, Wang YX, Hua F et al: Organization of NMDA receptors at extrasynaptic locations. Neuroscience 2010; 167: 68–87.
Wang CY, Chang K, Petralia RS, Wang YX, Seabold GK, Wenthold RJ : A novel family of adhesion-like molecules that interacts with the NMDA receptor. J Neurosci 2006; 26: 2174–2183.
Petralia RS, Wang Y-X, Wenthold RJ : NMDA receptors and PSD-95 are found in attachment plaques in cerebellar granular layer glomeruli. Eur J Neurosci 2002; 15: 583–587.
Traynelis SF, Wollmuth LP, McBain CJ et al: Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev 2010; 62: 405–496.
Karlsgodt KH, Robleto K, Trantham-Davidson H et al: Reduced dysbindin expression mediates N-methyl-D-aspartate receptor hypofunction and impaired working memory performance. Biol Psychiatry 2011; 69: 28–34.
Milenkovic M, Mielnik CA, Ramsey AJ : NMDA receptor-deficient mice display sexual dimorphism in the onset and severity of behavioural abnormalities. Genes Brain Behav 2014; 13: 850–862.
Driesen NR, McCarthy G, Bhagwagar Z et al: The impact of NMDA receptor blockade on human working memory-related prefrontal function and connectivity. Neuropsychopharmacology 2013; 38: 2613–2622.
Collette F, Hogge M, Salmon E, Van der Linden M : Exploration of the neural substrates of executive functioning by functional neuroimaging. Neuroscience 2006; 139: 209–221.
Ando J, Ono Y, Wright MJ : Genetic structure of spatial and verbal working memory. Behav Genet 2001; 31: 615–624.
Chen L-S, Rice TK, Thompson PA, Barch DM, Csernansky JG : Familial aggregation of clinical and neurocognitive features in sibling pairs with and without schizophrenia. Schizophr Res 2009; 111: 159–166.
Kang C, Drayna D : Genetics of speech and language disorders. Annu Rev Genom Hum Genet 2011; 12: 145–164.
Thevenon J, Callier P, Andrieux J et al: 12p13.33 microdeletion including ELKS/ERC1, a new locus associated with childhood apraxia of speech. Eur J Hum Genet 2013; 21: 82–88.
Baddeley A, Jarrold C : Working memory and Down syndrome. J Intellect Disabil Res 2007; 51: 925–931.
Carney DPJ, Brown JH, Henry LA : Executive function in Williams and Down syndromes. Res Dev Disabil 2013; 34: 46–55.
Arias-Vásquez A, Altink ME, Rommelse NNJ et al: CDH13 is associated with working memory performance in attention deficit/hyperactivity disorder. Genes Brain Behav 2011; 10: 844–851.
Bates TC, Luciano M, Medland SE, Montgomery GW, Wright MJ, Martin NG : Genetic variance in a component of the language acquisition device: ROBO1 polymorphisms associated with phonological buffer deficits. Behav Genet 2011; 41: 50–57.
Kaufman L, Ayub M, Vincent JB : The genetic basis of non-syndromic intellectual disability: a review. J Neurodev Disord 2010; 2: 182–209.
Nithianantharajah J, Komiyama NH, McKechanie A et al: Synaptic scaffold evolution generated components of vertebrate cognitive complexity. Nat Neurosci 2013; 16: 16–24.
Dolan J, Walshe K, Alsbury S et al: The extracellular leucine-rich repeat superfamily; a comparative survey and analysis of evolutionary relationships and expression patterns. BMC Genom 2007; 8: 320.
Thaler A, Mirelman A, Gurevich T et al: LRRK2 Ashkenazi Jewish Consortium. Lower cognitive performance in healthy G2019S LRRK2 mutation carriers. Neurology 2012; 79: 1027–1032.
De Bruijn DRH, van Dijk AHA, Pfundt R et al: Severe progressive autism associated with two de novo changes: a 2.6-Mb 2q31.1 deletion and a balanced t(14;21)(q21.1;p11.2) translocation with long-range epigenetic silencing of LRFN5 expression. Mol Syndr 2010; 1: 46–57.
Mikhail FM, Lose EJ, Robin NH et al: Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet A 2011; 155A: 2386–2396.
Kleffmann W, Zink AM, Lee JA et al: 5q31 Microdeletions: definition of a critical region and analysis of LRRTM2, a candidate gene for intellectual disability. Mol Syndr 2012; 3: 68–75.
Sousa I, Clark TG, Holt R et al: International Molecular Genetic Study of Autism Consortium (IMGSAC). Polymorphisms in leucine-rich repeat genes are associated with autism spectrum disorder susceptibility in populations of European ancestry. Mol Autism 2010; 1: 7.
Carlisle HJ, Luong TN, Medina-Marino A et al: Deletion of densin-180 results in abnormal behaviors associated with mental illness and reduces mGluR5 and DISC1 in the postsynaptic density fraction. J Neurosci Off J. Soc Neurosci 2011; 31: 16194–16207.
Katayama K, Yamada K, Ornthanalai VG et al: Slitrk1-deficient mice display elevated anxiety-like behavior and noradrenergic abnormalities. Mol Psychiatry 2010; 15: 177–184.
Takahashi H, Katayama K-I, Sohya K et al: Selective control of inhibitory synapse development by Slitrk3-PTPδ trans-synaptic interaction. Nat Neurosci 2012; 15: 389–398.
Takashima N, Odaka YS, Sakoori K et al: Impaired cognitive function and altered hippocampal synapse morphology in mice lacking Lrrtm1, a gene associated with schizophrenia. PLoS One 2011; 6: e22716.
Seabold GK, Wang PY, Petralia RS et al: Dileucine and PDZ-binding motifs mediate synaptic adhesion-like molecule 1 (SALM1) trafficking in hippocampal neurons. J Biol Chem 2012; 287: 4470–4484.
Ko J, Kim S, Chung HS et al: SALM synaptic cell adhesion-like molecules regulate the differentiation of excitatory synapses. Neuron 2006; 50: 233–245.
Mah W, Ko J, Nam J, Han K, Chung WS, Kim E : Selected SALM (synaptic adhesion-like molecule) family proteins regulate synapse formation. J Neurosci 2010; 30: 5559–5568.
Morimura N, Inoue T, Katayama K, Aruga J : Comparative analysis of structure, expression and PSD95-binding capacity of Lrfn, a novel family of neuronal transmembrane proteins. Gene 2006; 380: 72–83.
Wang SS, Kloth AD, Badura A : The cerebellum, sensitive periods, and autism. Neuron 2014; 83: 518–532.
Voineagu I, Wang X, Johnston P et al: Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 2011; 474: 380–384.
Acknowledgements
We thank the family for their participation in this study. We also thank the regional council of Burgundy for the financial support of the project. GKS, YW, and RSP were supported by the NIDCD/NIH Intramural Research Program.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Thevenon, J., Souchay, C., Seabold, G. et al. Heterozygous deletion of the LRFN2 gene is associated with working memory deficits. Eur J Hum Genet 24, 911–918 (2016). https://doi.org/10.1038/ejhg.2015.221
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ejhg.2015.221
This article is cited by
-
Genome-wide association study for multiple phenotype analysis
BMC Proceedings (2018)
-
High-fidelity CRISPR/Cas9- based gene-specific hydroxymethylation rescues gene expression and attenuates renal fibrosis
Nature Communications (2018)
-
Autism-like behaviours and enhanced memory formation and synaptic plasticity in Lrfn2/SALM1-deficient mice
Nature Communications (2017)