Abstract
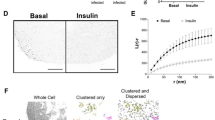
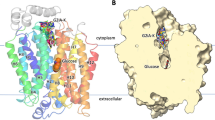
Cellular uptake of glucose is a tightly controlled process. It is mediated by a family of intrinsic membrane proteins (facilitated glucose transporters or GLUTs), and further regulated by metabolites and hormones. By far the most important GLUT regulation in human physiology is that by insulin, dysfunction of which may produce insulin resistance, a hallmark of diabetes mellitus and obesity. Six isoforms are now known in GLUT family. They differ in tissue-specific expression and regulation, yet share a common transmembrane topology showing a highly conserved transmembrane domain, and less conserved cytoplasmic and exoplasmic domains. Evidence indicates that the transmembrane domain accommodates a waterfilled glucose pathway (the catalytic domain). Little is known about the role of the exoplasmic domain. As for the cytoplasmic domain, a large body of evidence indicates its importance in tissue-specific regulation of GLUT function (the regulatory domain). This domain includes the N-terminal segment, the large, central loop, and the Cterminal segment. Exactly how this domain participates in GLUT regulation is not known. An interesting possibility is that the regulation involves a specific cellular protein, which interacts with GLUT at the cytoplasmic domain and modulates the function. This putative, glucose transporter binding protein (GTBP) may be an enzyme, or a nonenzymatic adaptor or docking protein. Indeed, we have identified several cytosolic proteins that bind to the cytoplasmic domain of GLUT proteins; these include glyceraldehyde-3-phosphate dehydrogenase, glucokinase, GTBP70, GTBP85, GTBP28, and L- 3 - hydroxyacyl CoA dehydrogenase. Interaction of GLUT protein with some of these GTPBs are functionally coupled. Whether any of these interactions is actually responsible for the insulin-induced GLUT regulation is yet to be determined.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Jung, C. The facilitative glucose transporter and insulin action. Exp Mol Med 28, 153–160 (1996). https://doi.org/10.1038/emm.1996.24
Published:
Issue date:
DOI: https://doi.org/10.1038/emm.1996.24
This article is cited by
-
Madhuca longifolia-hydro-ethanolic-fraction reverses mitochondrial dysfunction and modulates selective GLUT expression in diabetic mice fed with high fat diet
Molecular Biology Reports (2024)
-
High fat diet administration leads to the mitochondrial dysfunction and selectively alters the expression of class 1 GLUT protein in mice
Molecular Biology Reports (2019)