Abstract
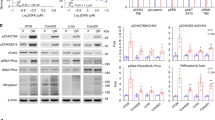
Checkpoint kinase 1 (Chk1) and Chk2 are effector kinases in the cellular DNA damage response and impairment of their function is closely related to tumorigenesis. Previous studies revealed several substrate proteins of Chk1 and Chk2, but identification of additional targets is still important in order to understand their tumor suppressor functions. In this study, we screened novel substrates for Chk1 and Chk2 using substrate target motifs determined previously by an oriented peptide library approach. The potential candidates were selected by genome-wide peptide database searches and were examined by in vitro kinase assays. ST5, HDAC5, PGC-1α, PP2A PR130, FANCG, GATA3, cyclin G, Rad51D and MAD1α were newly identified as in vitro substrates for Chk1 and/or Chk2. Among these, HDAC5 and PGC-1α were further analyzed to substantiate the screening results. Immunoprecipitation kinase assay of full-length proteins and site-directed mutagenesis analysis of the target motifs demonstrated that HDAC5 and PGC-1α were specific targets for Chk1 and/or Chk2 at least in vitro.
Similar content being viewed by others
Article PDF
Author information
Authors and Affiliations
Rights and permissions
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/3.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Kim, MA., Kim, HJ., Brown, A. et al. Identification of novel substrates for human checkpoint kinase Chk1 and Chk2 through genome-wide screening using a consensus Chk phosphorylation motif. Exp Mol Med 39, 205–212 (2007). https://doi.org/10.1038/emm.2007.23
Published:
Issue date:
DOI: https://doi.org/10.1038/emm.2007.23
Keywords
This article is cited by
-
GATA3 induces mitochondrial biogenesis in primary human CD4+ T cells during DNA damage
Nature Communications (2021)
-
UBR-box containing protein, UBR5, is over-expressed in human lung adenocarcinoma and is a potential therapeutic target
BMC Cancer (2020)
-
Centrosome-associated regulators of the G2/M checkpoint as targets for cancer therapy
Molecular Cancer (2009)
-
Akt2, but not Akt1, is required for cell survival by inhibiting activation of JNK and p38 after UV irradiation
Oncogene (2009)
-
The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men
Nature Reviews Molecular Cell Biology (2008)