Abstract
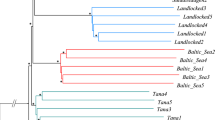
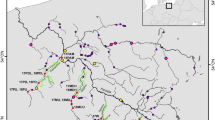
We describe genetic variation at three microsatellite single loci and six allozyme loci of seven natural Atlantic salmon populations from Ireland and Spain. A comparison of genetic variability detected at both types of loci is performed. Also, the relative value of microsatellite single locus variability with regard to the identification of individual salmon populations is assessed. Microsatellite loci are shown to display higher levels of variation than allozyme loci. Mean number of alleles (6 ± 1.53) and heterozygosity (0.46 ± 0.04) at microsatellite loci are greater than those found for allozymes (1.85 ±0.05 and 0.21 + 0.03, respectively), and some microsatellite alleles appear to be specific for a location or geographical area. Allozyme and microsatellite variation show the same pattern of differentiation between populations with Irish and Spanish populations grouped into different clusters. However, greater values of genetic distance were found among microsatellite (D = 0.0747 + 0.011) rather than among allozyme loci (D = 0.0449 + 0.008). These results provide evidence that microsatellite-based analysis of genetic variation will be useful in the identification of individual populations of Atlantic salmon.
Similar content being viewed by others
Article PDF
References
Aebersold, P B, Winans, G A, Teel, D J, Milner, G B, and Utter, F M. 1987. Manual for Starch Gel Electrophoresis: a Method for the Detection of Genetic Variation. Noaa Tech. Rep. NMFS. National Marine Fisheries Service. Seattle, WA.
Ayala, F J, and Powell, J R. 1972. Allozymes as diagnostic characters of sibling species of Drosophila. Proc Natl Acad Sci USA, 69, 1094–1096.
Blanco, G, Sánchez, J A, Vázquez, E, Rubio, J, and Utter, F M. 1992. Genetic differentiation among natural European populations of Atlantic salmon, Salmo salar L., from drainages of the Atlantic Ocean. Anim Genet, 23, 11–18.
Britten, R J, and Kohne, D. 1968. Repeated sequences in DNA. Science, 161, 529–540.
Clayton, J W, and Tretiak, D N. 1972. Amine-citrate buffers for pH control in starch gel electrophoresis. J Fish Res Board Can, 29, 1169–1172.
Cross, T F, and King, J. 1983. Genetic effects of hatchery rearing in Atlantic salmon. Aquaculture, 33, 33–40.
Cross, T F, and Ward, R D. 1980. Protein variation and duplicate loci in the Atlantic salmon, Salmo salar L. Genet Res, 36, 147–165.
Crozier, W W, and Moffet, I J J. 1989. Amount and distribution of biochemical and genetic variation among wild populations and hatchery stocks of Atlantic salmon, (Salmo salar L.), from north-east Ireland. J Fish Biol, 35, 665–677.
Davidson, W S, Birt, T P, and Green, J M. 1989. A review of genetic variation in Atlantic salmon, Salmo salar L., and its importance for stock identification, enhancement programmes and aquaculture. J Fish Biol, 34, 547–560.
Elo, K, Vourinen, J A, and Niemelä, E. 1994. Genetic resources of Atlantic salmon (Salmo salar L.) in Teno and Näätämö Rivers, northernmost Europe. Hereditas, 120, 19–28.
Estoup, A, Presa, P, Krieg, F, Vaiman, D, and Guyo-Mard, R. 1993. (CT)n and (GT)n microsatellites: a new class of genetic markers for Salmo trutta L. (brown trout). Heredity, 71, 488–496.
Galvin, P, Cross, T F, and Ferguson, A. 1994. Genetic differentiation and gene flow in Atlantic salmon, Salmo salar L.: a case study of the River Shannon system in Ireland. Aquacult Fish Manage, 25 (suppl. 2), 131–145.
Hamada, H, Petrino, M G, and Kakunaga, T. 1982. A novel repeated element with Z-DNA-forming potential is widely found in evolutionarily diverse eukaryotic genomes. Proc Natl Acad Sci USA, 79, 6465–6469.
Hearne, C M, Ghosh, S, and Todd, J A. 1992. Micro-satellites for linkage analysis of genetic traits. Trends Genet, 8, 288–294.
Hovey, S J, King, D P F, Thompson, D, and Scott, A. 1989. Mitochondrial DNA and allozyme analysis of Atlantic salmon, Salmo salar L., in England and Wales. J Fish Biol, 35, 253–260.
Hughes, C R, and Queller, D C. 1993. Detection of highly polymorphic microsatellite loci in species with little allozyme polymorphism. Mol Ecol, 2, 131–137.
Jordan, W C, Youngson, A F, Hay, D W, and Ferguson, A. 1992. Genetic protein variation in natural populations of Atlantic salmon (Salmo salar L.) in Scotland: temporal and spatial variation. Can J Fish Aquat Sci, 49, 1863–1872.
Kojonen, M-L. 1989. Electrophoretically detectable genetic variation in natural and hatchery stocks of Atlantic salmon in Finland. Hereditas, 110, 23–35.
Kreitman, M. 1983. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature, 304, 412–417.
McElligott, E A, and Cross, T F. 1991. Protein variation in wild Atlantic salmon, with particular reference to southern Ireland. J Fish Biol, 39, 35–42.
Mooney, J, Powell, E, Clabby, C, and Powell, R. 1995. Detection of Aeromonas salmonicida in wild Atlantic salmon using a specific DNA probe test. Dis aquat Org, 21, 131–135.
Nei, M. 1972. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA, 70, 3321–3323.
O'Connell, M, Skibinsky, D O F, and Beardmore, J A. 1995. Mitochondrial DNA and allozyme variation in Atlantic salmon (Salmo salar) populations in Wales. Can J Fish Aquat Sci, 52, 171–178.
Payne, R F L, Child, A R, and Forrest, A. 1971. Geographical variation in the Atlantic salmon. Nature, 231, 250–252.
Presa, P, Krieg, F, Estoup, A, and Guyomard, R. 1994. Diversité et gestion génétique de la truite commune: apport de l'étude du polymorphisme des locus protéiques et microsatellites. Génét Séi Évol, 26, 183–202.
Ridgway, G J, Sherburne, S W, and Lewis, R D. 1970. Polymorphism in esterases of Atlantic herring. Trans Am Fish Soc, 99, 147–151.
Sánchez, J A, Blanco, G, and Vázquez, E. 1993. Genetic status of Atlantic salmon in Asturias (northern Spain). In: Cloud, J. G. and Thorgaard, G. H. (eds) Genetic Conservation of Salmonid Fishes, pp. 219–225. Plenum Press, New York.
Sánchez, J A, Blanco, G, Vázquez, E, García, E, and Rubio, J. 1991. Allozyme variation in natural populations of Atlantic salmon in Asturias (northern Spain). Aquaculture, 93, 291–298.
Sanger, F, Nicklen, S, and Coulson, A R. 1977. DNA sequencing with chain terminating inhibitors. Proc Natl Acad Sci USA, 77, 5463–5467.
Shaklee, J B, Allendorf, F W, Morizot, D C, and Whitt, G S. 1990. Gene nomenclature for protein-coding loci in fish. Trans Am Fish Soc, 119, 2–15.
Slettan, A, Olsaker, I, and Lie, O. 1993. Isolation and characterization of variable (GT)n repetitive sequences from Atlantic salmon, Salmo salar L. Anim Genet, 24, 195–197.
Sneath, P H A, and Sokal, R R. 1973. Numerical Taxonomy. W.H. Freeman, San Francisco, CA.
Spratt, B G, Hedge, P J, Heesesen, S, Edelman, A, and Broome-Smith, J K. 1986. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene, 41, 337–342.
Stähl, G. 1987. Genetic population structure of Atlantic salmon. In: Ryman, N. and Utter, F., (eds) Population Genetics and Fishery Management, pp. 121–140. University of Washington Press, Seattle.
Swofford, D L, and Selander, R B. 1989. BIOSYS-I: a FORTRAN program for the comprehensive analysis of electrophoretic data in population genetics and systematics. J Hered, 72, 282–302.
Taggart, J B, and Ferguson, A. 1990. Hypervariable minisatellite DNA single locus probes for the Atlantic salmon, Salmo salar L. J Fish Biol, 37, 991–993.
Verspoor, E. 1988. Identification of stocks in the Atlantic salmon. In: Stroud, R. H. (ed.) Proceedings of the Symposium on Future Atlantic Salmon Management, pp. 37–46. Marine Recreational Fisheries Series, Savannah, GA.
Verspoor, E, and Jordan, W C. 1989. Genetic variation at the Me-2 locus in the Atlantic salmon within and between rivers: evidence for selective maintenance. J Fish Biol, 35, 205–213.
Verspoor, E, and Jordan, W C. 1994. Detection of an NAD+-dependent malic enzyme locus in the Atlantic salmon, Salmo salar, and other salmonid fish. Biochem Genet, 32, 105–117.
Weber, J L. 1990. Informativeness of human (dC-dA)n (dG-dT)n polymorphism. Genomics, 7, 524–530.
Wilson, I F, Bourke, E A, and Cross, T F. 1995. A triose-phosphate isomerase polymorphism in the Atlantic salmon, Salmo salar L. Biochem Genet, 33, 25–33.
Wintero, A K, Fredholm, M, and Thomsen, P D. 1992. Variable (dG-dT)n, (dC-dA)n sequences in the porcine genome. Genomics, 12, 281–288.
Wong, A K C, Yee, H A, Van De Sande, J H, and Rattner, J B. 1990. Distribution of CT-rich tracts is conserved in vertebrate chromosome. Chromosoma, 99, 344–351.
Wright, S. 1978. Evolution and the Genetics of Populations, vol. 4, Variability Within and Among Natural Populations. University of Chicago, Chicago.
Acknowledgements
Acknowledgements This work was supported by the European Union FAR Programme (AQ-2-493) and Spanish Government (DIGICYT PB 90-0992). For collection of Irish fish, we thank the staff of the Central, Western and Eastern Fisheries Boards (particularly P. Gargan and W. Roche). Also, W. O'Connor and the staff of the Fisheries Conservation Unit, Hydro Generation Group, Electricity Supply Board. C. Clabby is supported by Forbairt and UCG postgraduate fellowships and D. Ramos by a FICYT postgraduate fellowship.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Sánchez, J., Clabby, C., Ramos, D. et al. Protein and microsatellite single locus variability in Salmo salar L. (Atlantic salmon). Heredity 77, 423–432 (1996). https://doi.org/10.1038/hdy.1996.162
Received:
Issue date:
DOI: https://doi.org/10.1038/hdy.1996.162
Keywords
This article is cited by
-
Chromosome aberrations in pressure-induced triploid Atlantic salmon
BMC Genetics (2020)
-
Production and verification of the first Atlantic salmon (Salmo salar L.) clonal lines
BMC Genetics (2020)
-
Epistatic regulation of growth in Atlantic salmon revealed: a QTL study performed on the domesticated-wild interface
BMC Genetics (2020)
-
Parentage assignment in Salmo trutta strains and their crossbreeds with known mating
Environmental Biology of Fishes (2020)
-
Implications for introgression: has selection for fast growth altered the size threshold for precocious male maturation in domesticated Atlantic salmon?
BMC Evolutionary Biology (2018)