Abstract
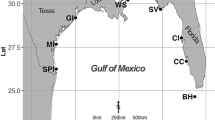
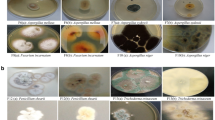
To elucidate fungal diversity in methane hydrate-bearing deep-sea marine sediments in the South China Sea, internal transcribed spacer (ITS) regions of rRNA genes from five different sediment DNA samples were amplified and phylogenetically analyzed. Total five ITS libraries were constructed and 413 clones selected randomly were grouped into 24 restriction patterns by Amplified Ribosomal DNA Restriction Analysis (ARDRA). ITS sequences of 44 representative clones were determined and compared with the GenBank database using gapped-BLAST. The phylogenetic analysis showed that the ITS sequences (71–97% similarity) were similar to those of Phoma, Lodderomyces, Malassezia, Cryptococcus, Cylindrocarpon, Hortaea, Pichia, Aspergillus and Candida. The remaining sequences were not associated to any known fungi or fungal sequences in the public database. The results suggested that methane hydrate-bearing deep-sea marine sediments harbor diverse fungi. This is the first report on fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Anderson IC, Cairney JWG . (2004). Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ Microbiol 6: 769–779.
Anderson IC, Campbell CD, Prosser JI . (2003a). Diversity of fungi in organic soils under a moorland-Scots pine (Pinus sylvestris L.) gradient. Environ Microbiol 5: 1121–1132.
Anderson IC, Campbell CD, Prosser JI . (2003b). Potential bias of fungal 18S rDNA and internal transcribed spacer polymerase chain reaction primers for estimating fungal biodiversity in soil. Environ Microbiol 5: 36–47.
Altschul SF, Madden TL, Schaffer AA, Zhang JH, Miller W, Lipman D . (1997). Gapped-BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
Baross JA, Hanus FJ, Morita RY . (1975). Survival of human enteric and other sewage microorganisms under simulated deep-sea conditions. Appl Microbiol 30: 309–318.
Buchan A, Newell SY, Moretam JIL, Moranm MA . (2002). Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern US salt marsh. Microb Ecol 43: 329–340.
Chen DM, Cairney JWG . (2002). Investigation of the influence of prescribed burning on ITS profiles of ectomycorrhizal and other soil fungi at three Australian sclerophyll forest sites. Mycol Res 106: 532–540.
Gabor EM, Vries EJ, Janssen DB . (2003). Efficient recovery of environmental DNA for expression cloning by indirect extraction methods. FEMS Microbiol Ecol 44: 153–163.
Gardes M, Bruns TD . (1993). ITS primers with enhanced specificity for basidiomycetes: application to the identification of mycorrhiza and rusts. Mol Ecol 2: 113–118.
Gellissen G . (2000). Heterologous protein production in methylotrophic yeasts. Appl Microbiol Biotechnol 54: 741–750.
Good IJ . (1953). The population frequencies of species and the estimation of the population parameters. Biometrika 40: 237–264.
Gotelli NJ, Entsminger GL . (2005). EcoSim: null models software for ecology. Version 7. Acquired Intelligence Inc. & Kesey-Bear. Jericho, VT 05465.http://garyentsminger.com/ecosim.htm.
Kohlmeyer J, Volkmann-Kohlmeyer B . (1991). Illustrated key to the filamentous higher marine fungi. Bot Mar 34: 1–61.
Liu XW, Li MF, Zhang YW, Zhang GX, Wu NY, Huang YY et al. (2005). Studies of seismic characteristics about gas hydrate: A case study of line HD152 in the South China Sea. Geoscience 19: 33–38 (in Chinese).
López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D . (2001). Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409: 603–606.
MacDonald IR, Guinasso NL, Sassen R, Brooks JM, Lee L, Scott KT . (1994). Gas hydrate that breaches the sea-floor on the continental-slope of the Gulf of Mexico. Geology 22: 699–702.
Maidak BL, Cole JR, Lilburn TG, Parker CY, Saxman PR, Farris RJ et al. (2001). The RDP-α (ribosomal database project). Nucleic Acids Res 29: 173–174.
Miller DN . (2001). Evaluation of gel filtration resins for the removal of PCR-inhibitory substances from soils and sediments. J Microbiol Meth 44: 48–58.
Mills HJ, Martinez RJ, Story S, Sobecky PA . (2005). Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl Environ Microbiol 71: 3235–3247.
Moon-van der Staay SY, De Wachter R, Vaulot D . (2001). Oceanic 18SrDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409: 607–610.
Nagahama T, Hamamoto M, Nakase T, Takaki Y, Horikoshi K . (2003). Cryptococus surugaensis sp. nov., a novel yeast species from sediment collected on the deep-sea floor of Suruga Bay. Int J Syst Evol Microbiol 53: 2095–2098.
Orphan VJ, House CH, Hinrichs KU, Mckeegan KD, Delong EF . (2002). Mutiple archaeal groups mediate methane oxidation in anoxic cold seep sediments. Proc Natl Acad Sci USA 99: 7663–7668.
Pivkin MV . (2000). Filamentous fungi associated with holothurians from the sea of Japan, off the primorye coast of Russia. Biol Bull 198: 101–109.
Rappé MS, Suzuki MT, Vergin KL, Giovannoni SJ . (1998). Phylogenetic diversity of ultraplankton plastid small-subunit rRNA genes recovered in environmental nucleic acid samples from the Pacific and Atlantic coasts of the United States. Appl Environ Microbiol 64: 294–303.
Reed DW, Fujita Y, Delwiche ME, Blackwelder DB, Sheridan PP, Uchida T et al. (2002). Microbial communities from methane hydrate-bearing deep marine sediments in a forearc basin. Appl Environ Microbiol 68: 3759–3770.
Robe P, Nalin R, Capellano C, Vogel TM, Simonet P . (2003). Extraction of DNA from soil. Eur J Soil Biol 39: 183–190.
Sassen R, Joye S, Sweet ST, DeFreitas DA, Milkov AV, MacDonald IR . (1999). Thermogenic gas hydrates and hydrocarbon gases in comples chemosynthetic communities, Gulf of Mexcio continental slope. Org Geochem 30: 485–497.
Shannon CE, Weaver W . (1949). The Mathematical Theory of Communication. University of Illinois Press: Urbana, USA.
Shao Z, Sun F . (2007). Intracellular sequestration of manganese and phosphorous in a metal-resistant fungus Cladosporium cladosporioides from deep-sea sediment. Extremophiles 11: 435–443.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored ‘rare biosphere’. Proc Natl Acad Sci USA 103: 12115–12120.
Stocek T, Epstein S . (2003). Novel eukaryotic lineages inferred from small-subunit rRNA analyses of oxygen-depleted marine environments. Appl Environ Microbiol 69: 2657–2663.
Stocek T, Taylor GT, Epstein S . (2003). Novel eukaryotes from the permanently anoxic Cariaco Basin (Caribbean Sea). Appl Environ Microbiol 69: 5656–5663.
Takami H, Inoue A, Horikoshi K . (1997). Microbial flora in the deepest sea mud of the Mariana Trench. FEMS Microbiol Lett 152: 279–285.
Takishita K, Tsuchiya M, Reimer JD, Maruyama T . (2006). Molecular evidence demonstrating the basidiomycetous fungus Cryptococcus curvatus is the dominant microbial eukaryote in sediment at the Kuroshima Knoll methane seep. Extremophiles 10: 165–169.
Viaud M, Pasquier A, Brygoo Y . (2000). Diversity of soil fungi studied by PCR-RFLP of ITS. Mycol Res 104: 1027–1032.
White TJ, Bruns TD, Lee S, Taylor J . (1990). Analysis of phylogenetic relationships by amplification and direct sequencing of ribosomal RNA genes. In: Innis MA, Gelfand DH, Sninsky JJ, White J (eds). PCR Protocols: a Guide to Methods and Applications. Academic Press: New York, pp 315–322.
Yan T, Ye Q, Zhou J, Zhang CL . (2006). Diversity of functional genes for methanotrophs in sediments associated with gas hydrates and hydrocarbon seeps in the Gulf of Mexico. FEMS Microbiol Ecol 57: 251–259.
Zhang GX, Huang YY, Zhu YH, Wu BH . (2002). Prospect of gas hydrate resources in the South China Sea. Mar Geol Tern Geol 22: 75–81 (in Chinese).
Acknowledgements
This work was supported by grant from the National High Technology Research and Development Plan of China (2003AA620401). We thank Yongyang Huang and Jian Liu, Guangzhou Marine Geological Survey, for their assistance during this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lai, X., Cao, L., Tan, H. et al. Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J 1, 756–762 (2007). https://doi.org/10.1038/ismej.2007.51
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2007.51
Keywords
This article is cited by
-
Sedimentary DNA for tracking the long-term changes in biodiversity
Environmental Science and Pollution Research (2023)
-
Adaptation mechanisms of the soil microbial community under stoichiometric imbalances and nutrient-limiting conditions in a subtropical nitrogen-saturated forest
Plant and Soil (2023)
-
Assessment of yeasts in tropical peat swamp forests in Thailand
Mycological Progress (2020)
-
Cultivable fungi present in deep-sea sediments of Antarctica: taxonomy, diversity, and bioprospecting of bioactive compounds
Extremophiles (2020)
-
Characterization of the total and viable bacterial and fungal communities associated with the International Space Station surfaces
Microbiome (2019)