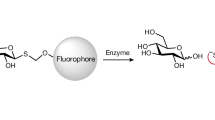
Abstract
In this study, the mining of an Antarctic soil sample by functional metagenomics allowed the isolation of a cold-adapted protein (RBcel1) that hydrolyzes only carboxymethyl cellulose. The new enzyme is related to family 5 of the glycosyl hydrolase (GH5) protein from Pseudomonas stutzeri (Pst_2494) and does not possess a carbohydrate-binding domain. The protein was produced and purified to homogeneity. RBcel1 displayed an endoglucanase activity, producing cellobiose and cellotriose, using carboxymethyl cellulose as a substrate. Moreover, the study of pH and the thermal dependence of the hydrolytic activity shows that RBcel1 was active from pH 6 to pH 9 and remained significantly active when temperature decreased (18% of activity at 10 °C). It is interesting that RBcel1 was able to synthetize non-reticulated cellulose using cellobiose as a substrate. Moreover, by a combination of bioinformatics and enzyme analysis, the physiological relevance of the RBcel1 protein and its mesophilic homologous Pst_2494 protein from P. stutzeri, A1501, was established as the key enzymes involved in the production of cellulose by bacteria. In addition, RBcel1 and Pst_2494 are the two primary enzymes belonging to the GH5 family involved in this process.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Almin KE, Eriksson KE . (1967). Enzymic degradation of polymers. I. Viscometric method for the determination of enzymic activity. Biochim Biophys Acta 139: 238–247.
Amann RI, Ludwig W, Schleifer KH . (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169.
Asgeirsson B, Nielsen BN, Højrup P . (2003). Amino acid sequence of the cold-active alkaline phosphatase from Atlantic cod (Gadus morhua). Comp Biochem Physiol B Biochem Mol Biol 136: 45–60.
Aurilia V, Parracino A, Saviano M, Rossi M, D'Auria S . (2007). The psychrophilic bacterium Pseudoalteromonas halosplanktis TAC125 possesses a gene coding for a cold-adapted feruloyl esterase activity that shares homology with esterase enzymes from gamma-proteobacteria and yeast. Gene 397: 51–57.
Benko Z, Drahos E, Szengyel Z, Puranen T, Vehmaanperä J, Réczey K . (2007). Thermoascus aurantiacus CBHI/Cel7A production in Trichoderma reesei on alternative carbon sources. Appl Biochem Biotechnol 137–140: 195–204.
Branda SS, Vik S, Friedman L, Kolter R . (2005). Biofilms: the matrix revisited. Trends Microbiol 13: 20–26.
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B . (2009). The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res 37 (Database issue): D233–D238.
Collins T, D'Amico S, Georlette D, Marx JC, Huston AL, Feller G . (2006). A nondetergent sulfobetaine prevents protein aggregation in microcalorimetric studies. Anal Biochem 352: 299–301.
Collins T, De Vos D, Hoyoux A, Savvides SN, Gerday C, Van Beeumen J et al. (2005). Study of the active site residues of a glycoside hydrolase family 8 xylanase. J Mol Biol 354: 425–435.
Da Re S, Ghigo JM . (2006). A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J Bacteriol 188: 3073–3087.
Davey ME, O'Toole GA . (2000). Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64: 847–867.
Delille D, Coulon F, Pelletier E . (2004). Biostimulation of natural microbial assemblages in oil-amended vegetated and desert sub-Antarctic soils. Microb Ecol 47: 407–415.
Delmer DP . (1999). Cellulose biosynthesis: exciting times for a difficult field of study. Annu Rev Plant Physiol Plant Mol Biol 50: 245–276.
Duchaud E, Boussaha M, Loux V, Bernardet JF, Michel C, Kerouault B et al. (2007). Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat Biotechnol 25: 763–769.
Feller G, d'Amico D, Gerday C . (1999). Thermodynamic stability of a cold-active alpha-amylase from the Antarctic bacterium. Alteromonas haloplanctis. Biochemistry 38: 4613–4619.
Feng Y, Duan CJ, Pang H, Mo XC, Wu CF, Yu Y et al. (2007). Cloning and identification of novel cellulase genes from uncultured microorganisms in rabbit cecum and characterization of the expressed cellulases. Appl Microbiol Biotechnol 75: 319–328.
Garsoux G, Lamotte J, Gerday C, Feller G . (2004). Kinetic and structural optimization to catalysis at low temperatures in a psychrophilic cellulase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Biochem J 384 (Part 2): 247–253.
Gerday C, Aittaleb M, Bentahir M, Chessa JP, Claverie P, Collins T et al. (2000). Cold-adapted enzymes: from fundamentals to biotechnology. Trends Biotechnol 18: 103–107.
Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR et al. (2002). Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol 68: 4301–4306.
Handelsman J . (2004). Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68: 669–685.
Hardeman F, Sjoling S . (2007). Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol Ecol 59: 524–534.
Henne A, Daniel R, Schmitz RA, Gottschalk G . (1999). Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol 65: 3901–3907.
Hong J, Tamaki H, Yamamoto K, Kumagai H . (2003). Cloning of a gene encoding a thermo-stable endo-beta-1,4-glucanase from Thermoascus aurantiacus and its expression in yeast. Biotechnol Lett 25: 657–661.
Ito S, Kobayashi T, Hatada Y, Horikoshi K . (2005). Enzymes in Modern Detergents. Methods Biotechnol 17: 151–163.
Lo Leggio L, Parry NJ, Van Beeumen J, Claeyssens M, Bhat MK, Pickersgill RW . (1997). Crystallization and preliminary X-ray analysis of the major endoglucanase from Thermoascus aurantiacus. Acta Crystallogr D Biol Crystallogr 53 (Part 5): 599–604.
Lonhienne T, Baise E, Feller G, Bouriotis V, Gerday C . (2001). Enzyme activity determination on macromolecular substrates by isothermal titration calorimetry: application to mesophilic and psychrophilic chitinases. Biochim Biophys Acta 1545: 349–356.
Matthysse AG, Deora R, Mishra M, Torres AG . (2008). Polysaccharides cellulose, poly-beta-1,6-n-acetyl-D-glucosamine, and colanic acid are required for optimal binding of Escherichia coli O157:H7 strains to alfalfa sprouts and K-12 strains to plastic but not for binding to epithelial cells. Appl Environ Microbiol 74: 2384–2390.
Matthysse AG, Marry M, Krall L, Kaye M, Ramey BE, Fuqua C et al. (2005). The effect of cellulose overproduction on binding and biofilm formation on roots by Agrobacterium tumefaciens. Mol Plant Microbe Interact 18: 1002–1010.
Matthysse AG, McMahan S . (1998). Root colonization by Agrobacterium tumefaciens is reduced in cel, attB, attD, and attR mutants. Appl Environ Microbiol 64: 2341–2345.
Matthysse AG, Thomas DL, White AR . (1995). Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J Bacteriol 177: 1076–1081.
Médigue C, Krin E, Pascal G, Barbe V, Bernsel A, Bertin PN et al. (2005). Coping with cold: the genome of the versatile marine Antarctica bacterium Pseudoalteromonas haloplanktis TAC125. Genome Res 15: 1325–1335.
Methé BA, Nelson KE, Deming JW, Momen B, Melamud E, Zhang X et al. (2005). The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA 102: 10913–10918.
Narinx E, Baise E, Gerday C . (1997). Subtilisin from psychrophilic Antarctic bacteria: characterization and site-directed mutagenesis of residues possibly involved in the adaptation to cold. Protein Eng 10: 1271–1279.
Parry NJ, Beever DE, Owen E, Nerinckx W, Claeyssens M, Van Beeumen J et al. (2002). Biochemical characterization and mode of action of a thermostable endoglucanase purified from Thermoascus aurantiacus. Arch Biochem Biophys 404: 243–253.
Rabus R, Ruepp A, Frickey T, Rattei T, Fartmann B, Stark M et al. (2004). The genome of Desulfotalea psychrophila, a sulfate-reducing bacterium from permanently cold Arctic sediments. Environ Microbiol 6: 887–902.
Ranjan R, Grover A, Kapardar RK, Sharma R . (2005). Isolation of novel lipolytic genes from uncultured bacteria of pond water. Biochem Biophys Res Commun 335: 57–65.
Rappe MS, Giovannoni SJ . (2003). The uncultured microbial majority. Annu Rev Microbiol 57: 369–394.
Recouvreux DO, Carminatti CA, Pitlovanciv AK, Rambo CR, Porto LM, Antônio RV . (2008). Cellulose biosynthesis by the beta-proteobacterium, Chromobacterium violaceum. Curr Microbiol 57: 469–476.
Rhee JK, Ahn DG, Kim YG, Oh JW . (2005). New thermophilic and thermostable esterase with sequence similarity to the hormone-sensitive lipase family, cloned from a metagenomic library. Appl Environ Microbiol 71: 817–825.
Riesenfeld CS, Goodman RM, Handelsman J . (2004). Uncultured soil bacteria are a reservoir of new antibiotic resistance genes. Environ Microbiol 6: 981–989.
Rodríguez-Navarro DN, Dardanelli MS, Ruíz-Saínz JE . (2007). Attachment of bacteria to the roots of higher plants. FEMS Microbiol Lett 272: 127–136.
Romling U . (2002). Molecular biology of cellulose production in bacteria. Res Microbiol 153: 205–212.
Shepherd MG, Tong CC, Cole AL . (1981). Substrate specificity and mode of action of the cellulases from the thermophilic fungus Thermoascus aurantiacus. Biochem J 193: 67–74.
Smit G, Swart S, Lugtenberg BJ, Kijne JW . (1992). Molecular mechanisms of attachment of Rhizobium bacteria to plant roots. Mol Microbiol 6: 2897–2903.
Standal R, Iversen TG, Coucheron DH, Fjaervik E, Blatny JM, Valla S . (1994). A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the BCS operon. J Bacteriol 176: 665–672.
Steele NM, Sulová Z, Campbell P, Braam J, Farkas V, Fry SC . (2001). Ten isoenzymes of xyloglucan endotransglycosylase from plant cell walls select and cleave the donor substrate stochastically. Biochem J 355 (Part 3): 671–679.
Streit WR, Schmitz RA . (2004). Metagenomics–the key to the uncultured microbes. Curr Opin Microbiol 7: 492–498.
Tewari YB, Goldberg RN . (1989). Thermodynamics of hydrolysis of disaccharides. Cellobiose, gentiobiose, isomaltose, and maltose. J Biol Chem 264: 3966–3971.
Tirawongsaroj P, Sriprang R, Harnpicharnchai P, Thongaram T, Champreda V, Tanapongpipat S et al. (2008). Novel thermophilic and thermostable lipolytic enzymes from a Thailand hot spring metagenomic library. J Biotechnol 133: 42–49.
Ude S, Arnold DL, Moon CD, Timms-Wilson T, Spiers AJ . (2006). Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ Microbiol 8: 1997–2011.
Van Petegem F, Vandenberghe I, Bhat MK, Van Beeumen J . (2002). Atomic resolution structure of the major endoglucanase from Thermoascus aurantiacus. Biochem Biophys Res Commun 296: 161–166.
Wang LX . (2008). Chemoenzymatic synthesis of glycopeptides and glycoproteins through endoglycosidase-catalyzed transglycosylation. Carbohydr Res 343: 1509–1522.
Acknowledgements
We thank R Pirard, V Collard and C Jerome for their skilful technical assistance. PP is a member of the Carrera del Investigador Científico (CONICET), Argentina and a recipient of a Postdoctoral fellowship from the Belgian Science Policy Office (BELSPO). SD'A is a Belgian FNRS postdoctoral researcher. This work was supported by an FNRS Grant (Brussels, Belgium, FRFC, 2.4561.07).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Berlemont, R., Delsaute, M., Pipers, D. et al. Insights into bacterial cellulose biosynthesis by functional metagenomics on Antarctic soil samples. ISME J 3, 1070–1081 (2009). https://doi.org/10.1038/ismej.2009.48
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.48
Keywords
This article is cited by
-
Investigation of cold-active and mesophilic cellulases: opportunities awaited
Biomass Conversion and Biorefinery (2023)
-
Role of metagenomics in prospecting novel endoglucanases, accentuating functional metagenomics approach in second-generation biofuel production: a review
Biomass Conversion and Biorefinery (2023)
-
Experimental evidence for enzymatic cell wall dissolution in a microbial protoplast feeder (Orciraptor agilis, Viridiraptoridae)
BMC Biology (2022)
-
GeneHunt for rapid domain-specific annotation of glycoside hydrolases
Scientific Reports (2019)
-
Cold survival strategies for bacteria, recent advancement and potential industrial applications
Archives of Microbiology (2019)