Abstract
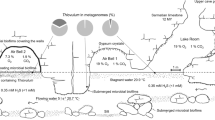
Microbial diversity in Movile Cave (Romania) was studied using bacterial and archaeal 16S rRNA gene sequence and functional gene analyses, including ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), soxB (sulfate thioesterase/thiohydrolase) and amoA (ammonia monooxygenase). Sulfur oxidizers from both Gammaproteobacteria and Betaproteobacteria were detected in 16S rRNA, soxB and RuBisCO gene libraries. DNA-based stable-isotope probing analyses using 13C-bicarbonate showed that Thiobacillus spp. were most active in assimilating CO2 and also implied that ammonia and nitrite oxidizers were active during incubations. Nitrosomonas spp. were detected in both 16S rRNA and amoA gene libraries from the ‘heavy’ DNA and sequences related to nitrite-oxidizing bacteria Nitrospira and Candidatus ‘Nitrotoga’ were also detected in the ‘heavy’ DNA, which suggests that ammonia/nitrite oxidation may be another major primary production process in this unique ecosystem. A significant number of sequences associated with known methylotrophs from the Betaproteobacteria were obtained, including Methylotenera, Methylophilus and Methylovorus, supporting the view that cycling of one-carbon compounds may be an important process within Movile Cave. Other sequences detected in the bacterial 16S rRNA clone library included Verrucomicrobia, Firmicutes, Bacteroidetes, alphaproteobacterial Rhodobacterales and gammaproteobacterial Xanthomonadales. Archaeal 16S rRNA sequences retrieved were restricted within two groups, namely the Deep-sea Hydrothermal Vent Euryarchaeota group and the Miscellaneous Crenarchaeotic group. No sequences related to known sulfur-oxidizing archaea, ammonia-oxidizing archaea, methanogens or anaerobic methane-oxidizing archaea were detected in this clone library. The results provided molecular biological evidence to support the hypothesis that Movile Cave is driven by chemolithoautotrophy, mainly through sulfur oxidation by sulfur-oxidizing bacteria and reveal that ammonia- and nitrite-oxidizing bacteria may also be major primary producers in Movile Cave.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E . (2007). Cultivation of a novel cold-adapted nitrite oxidizing Betaproteobacterium from the Siberian Arctic. ISME J 1: 256–264.
Chen Y, Dumont MG, Neufeld JD, Bodrossy L, Stralis-Pavese N, McNamara NP et al. (2008). Revealing the uncultured majority: combining DNA stable-isotope probing, multiple displacement amplification and metagenomics analyses of uncultivated Methylocystis in acidic peatlands. Environ Microbiol 10: 2609–2622.
Delong EF . (1992). Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685–5689.
Engel AS, Lee N, Porter ML, Stern LA, Bennett PC, Wagner M . (2003). Filamentous ‘Epsilonproteobacteria’ dominate microbial mats from sulfidic cave springs. Appl Environ Microbiol 69: 5503–5511.
Engel AS, Porter ML, Stern LA, Quinlan S, Bennett PC . (2004). Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic ‘Epsilonproteobacteria’. FEMS Microbiol Ecol 51: 31–53.
Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB . (2005). Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. PNAS 102: 14683–14688.
Galtier N, Gouy M, Gautier C . (1996). SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci 12: 543–548.
Giri BJ, Bano N, Hollibaugh JT . (2004). Distribution of RuBisCO genotypes along a redox gradient in Mono Lake, California. Appl Environ Microbiol 70: 3443–3448.
Hutchens E, Radajewski S, Dumont MG, McDonald IR, Murrell JC . (2004). Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ Microbiol 6: 111–120.
Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the sea of Okhotsk. Appl Environ Microbiol 69: 7224–7235.
Ito T, Sugita K, Yumoto I, Nodasaka Y, Okabe S . (2005). Thiovirga sulfuroxydans gen. nov., sp. nov., a chemolithoautotrophic sulfur-oxidizing bacterium isolated from a microaerobic waste-water biofilm. Int J Syst Evol Microbiol 55: 1059–1064.
Kelly DP, Shergill JK, Lu WP, Wood AP . (1997). Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71: 95–107.
Kodama Y, Watanabe K . (2004). Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int J Syst Evol Microbiol 54: 2297–2300.
Lam P, Cowen JP, Jones RD . (2004). Autotrophic ammonia oxidation in a deep-sea hydrothermal plume. FEMS Microbiol Ecol 47: 191–206.
Lane DJ . (1991). 16S/23S rRNA sequencing. In: Stackebrandt E and Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons: New York, USA, pp 115–147.
Limpiyakorn T, Kurisu F, Sakamoto Y, Yagi O . (2007). Effects of ammonium and nitrite on communities and populations of ammonia-oxidizing bacteria in laboratory-scale continuous-flow reactors. FEMS Microbiol Ecol 60: 501–512.
Lliros M, Casamayor EO, Borrego C . (2008). High archaeal richness in the water column of a freshwater sulfurous karstic lake along an interannual study. FEMS Microbiol Ecol 66: 331–342.
Ludwig W, Strunk O, Westram R, Richter L, Meier J, Yadhukumar H et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1372.
Macalady JL, Lyon EH, Koffman B, Albertson LK, Meyer K, Galdenzi S et al. (2006). Dominant microbial populations in limestone-corroding stream biofilms, Frasassi cave system, Italy. Appl Environ Microbiol 72: 5596–5609.
Macalady JL, Jones DS, Lyon EH . (2007). Extremely acidic, pendulous cave wall biofilms from the Frasassi cave system, Italy. Environ Microbiol 9: 1402–1414.
Macalady JL, Dattagupta S, Schaperdoth I, Jones DS, Druschel GK, Eastman D . (2008). Niche differentiation among sulfur-oxidizing bacterial populations in cave waters. ISME J 2: 590–601.
Meyer B, Imhoff JF, Kuever J . (2007). Molecular analysis of the distribution and phylogeny of the soxB gene among sulfur-oxidizing bacteria — evolution of the Sox sulfur oxidation enzyme system. Environ Microbiol 9: 2957–2977.
Mori K, Suzuki K . (2008). Thiofaba tepidiphila gen. nov., sp. nov., a novel obligate chemolithoautotrophic, sulfur-oxidizing bacterium of the Gammaproteobacteria isolated from a hot spring. Int J Syst Evol Microbiol 58: 1885–1891.
Muyzer G, de Waal EC, Uitterlinden AG . (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700.
Nakagawa S, Takai K . (2008). Deep-sea vent chemoautotrophs: diversity, biochemistry and ecological significance. FEMS Microbiol Ecol 65: 1–14.
Neufeld JD, Schafer H, Cox MJ, Boden R, McDonald IR, Murrell JC . (2007). Stable-isotope probing implicates Methylophaga spp. and novel Gammaproteobacteria in marine methanol and methylamine metabolism. ISME J 1: 480–491.
Otawa K, Asano R, Ohba Y, Sasaki T, Kawamura E, Koyama F et al. (2006). Molecular analysis of ammonia-oxidizing bacteria community in intermittent aeration sequencing batch reactors used for animal wastewater treatment. Environ Microbiol 8: 1985–1996.
Rohwerder T, Sand W, Lascu C . (2003). Preliminary evidence for a sulfur cycle in Movile Cave, Romania. Acta Biotechnol 23: 101–107.
Rotthauwe JH, Witzel KP, Liesack W . (1997). The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl Environ Microbiol 63: 4704–4712.
Sarbu SM, Kinkle BK, Vlasceanu L, Kane TC . (1994). Microbiological characterization of a sulfur-rich groundwater ecosystem. Geomicrobiol J 12: 175–182.
Sarbu SM, Kane TC . (1995). A subterranean chemoautotrophically based ecosystem. NSS Bull 57: 91–98.
Sarbu SM, Kane TC, Kinkle BK . (1996). A chemoautotrophically based cave ecosystem. Science 272: 1953–1955.
Sarbu SM . (2000). Movile Cave: a chemoautotrophically based groundwater ecosystem. In: Wilkens H, Culver DC, Humphreys WF (eds). Subterranean Ecosystems. Elsevier: Amsterdam, pp 319–343.
Selesi D, Schmid M, Hartmann A . (2005). Diversity of green-like and red-like ribulose-1,5-bisphosphate carboxylase/oxygenase large-subunit genes (cbbL) in differently managed agricultural soils. Appl Environ Microbiol 71: 175–184.
Takai K, Horikoshi K . (1999). Genetic diversity of Archaea in deep-sea hydrothermal vent environments. Genetics 152: 1285–1297.
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG . (1997). The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24: 4876–4882.
Vlasceanu L, Popa R, Kinkle BK . (1997). Characterization of Thiobacillus thioparus LV43 and its distribution in a chemoautotrophically based groundwater ecosystem. Appl Environ Microbiol 63: 3123–3127.
Acknowledgements
L Wu acknowledges the NERC International Opportunities Fund of the Marine & Freshwater Microbial Biodiversity Programme. Y Chen was supported by a NERC, Dorothy Hodgkin Postgraduate Award through the University of Warwick. We thank A Simpson, C Gibbs, L Witcomb, M Stein and M Bhatti for their help in construction of clone libraries. We greatly acknowledge Prof DP Kelly for his insight and advice on the paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, Y., Wu, L., Boden, R. et al. Life without light: microbial diversity and evidence of sulfur- and ammonium-based chemolithotrophy in Movile Cave. ISME J 3, 1093–1104 (2009). https://doi.org/10.1038/ismej.2009.57
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2009.57
Keywords
This article is cited by
-
A new extremophile ostracod crustacean from the Movile Cave sulfidic chemoautotrophic ecosystem in Romania
Scientific Reports (2023)
-
Environmental Drivers of the Moonmilk Microbiome Diversity in Some Temperate and Tropical Caves
Microbial Ecology (2023)
-
Metagenome Analysis of Speleothem Microbiome from Subterranean Cave Reveals Insight into Community Structure, Metabolic Potential, and BGCs Diversity
Current Microbiology (2023)
-
Competition-cooperation in the chemoautotrophic ecosystem of Movile Cave: first metagenomic approach on sediments
Environmental Microbiome (2022)
-
The diversity of microbes and prediction of their functions in karst caves under the influence of human tourism activities—a case study of Zhijin Cave in Southwest China
Environmental Science and Pollution Research (2022)