Abstract
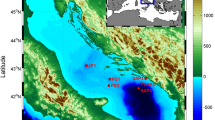
The ecosystems of the Red Sea are among the least-explored microbial habitats in the marine environment. In this study, we investigated the microbial communities in the water column overlying the Atlantis II Deep and Discovery Deep in the Red Sea. Taxonomic classification of pyrosequencing reads of the 16S rRNA gene amplicons showed vertical stratification of microbial diversity from the surface water to 1500 m below the surface. Significant differences in both bacterial and archaeal diversity were observed in the upper (2 and 50 m) and deeper layers (200 and 1500 m). There were no obvious differences in community structure at the same depth for the two sampling stations. The bacterial community in the upper layer was dominated by Cyanobacteria whereas the deeper layer harbored a large proportion of Proteobacteria. Among Archaea, Euryarchaeota, especially Halobacteriales, were dominant in the upper layer but diminished drastically in the deeper layer where Desulfurococcales belonging to Crenarchaeota became the dominant group. The results of our study indicate that the microbial communities sampled in this study are different from those identified in water column in other parts of the world. The depth-wise compositional variation in the microbial communities is attributable to their adaptations to the various environments in the Red Sea.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Amann RI, Ludwig W, Schleifer KH . (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169.
Anschutz P, Blanc G . (1996). Heat and salt fluxes in the Atlantis II Deep (Red Sea). Earth Planet Sci Lett 142: 147–159.
Baas-Becking LGM . (1934). Geobiologie of Inleiding Tot de Milieukunde. Van Stockkum & Zoon: The Hague, the Netherlands.
Bäcker H, Schoell M . (1972). New deeps with brines and metalliferous sediments in the Red Sea. Nature Phys Sci 240: 153–158.
Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH . (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci USA 105: 10583–10588.
Chao A . (1984). Nonparametric estimation of the number of classes in a population. Scand J Stat 11: 265–270.
Chao A, Ma MC, Yang MCK . (1993). Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80: 193–201.
Cho JC, Tiedje JM . (2000). Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl Environ Microbiol 66: 5448–5456.
Clarke KR (1993). Non-parametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143.
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ et al. (2009). The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37: D141–D145.
Cottrell MT, Kirchman DL . (2000). Community composition of marine bacterioplankton determined by 16S rRNA gene clone libraries and fluorescence in situ hybridization. Appl Environ Microbiol 66: 5116–5122.
Degens E, Ross D . (1969). Hot Brines and Recent Heavy Metal Deposits in the Red Sea. Springer: New York, NY.
DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU et al. (2006). Community genomics among stratified microbial assemblages in the ocean's interior. Science 311: 496–503.
Dick GJ, Lee YE, Tebo BM . (2006). Manganese(II)-oxidizing Bacillus spores in Guaymas basin hydrothermal sediments and plumes. Appl Environ Microbiol 72: 3184–3190.
Eder W, Jahnke LL, Schmidt M, Huber R . (2001). Microbial diversity of the brine-seawater interface of the Kebrit Deep, Red Sea, studied via 16S rRNA gene sequences and cultivation methods. Appl Environ Microbiol 67: 3077–3085.
Eder W, Schmidt M, Koch M, Garbe-Schönberg D, Huber R . (2002). Prokaryotic phylogenetic diversity and corresponding geochemical data of the brine–seawater interface of the Shaban Deep, Red Sea. Environ Microbiol 4: 758–763.
Edgar RC . (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797.
Eisen JA . (2007). Environmental shotgun sequencing: its potential and challenges for studying the hidden world of microbes. PLoS Biol 5: e82.
Good IJ . (1953). The population frequencies of species and the estimation of population parameters. Biometrika 40: 237–264.
Gurvich EG . (2006). Metalliferous sediments of the Red Sea. Metalliferous Sediments of the World Ocean. Springer: Berlin Heidelberg.
Hagstrom A, Pommier T, Rohwer F, Simu K, Stolte W et al. (2002). Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl Environ Microbiol 68: 3628–3633.
Hansen MC, Tolker-Nielsen T, Givskov M, Molin S . (1998). Biased 16S rDNA PCR amplification caused by interference from DNA flanking the template region. FEMS Microbiol Ecol 26: 141–149.
Hong S, Bunge J, Leslin C, Jeon S, Epstein SS . (2009). Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J 3: 1365–1373.
Huber H, Stetter KO . (2006). Desulfurococcales. The Prokaryotes vol. 3. Springer: New York.
Kendall MM, Boone DR . (2006). The Order Methanosarcinales. The Prokaryotes, vol. 3. Springer: New York.
Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E . (2006). Introduction to the Proteobacteria. The Prokaryotes vol. 5. Springer: New York.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380.
Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810.
Morris RM, Rappe MS, Urbach E, Connon SA, Giovannoni SJ . (2004). Prevalence of the Chloroflexi-related SAR202 bacterioplankton cluster throughout the mesopelagic zone and deep ocean. Appl Environ Microbiol 70: 2836–2842.
Neumann AC, Chave KE . (1965). Connate origin proposed for hot salty bottom water from a Red Sea basin. Nature 206: 1346–1347.
Oren A . (2006). The Order Halobacteriales. The Prokaryotes, vol. 3. Springer: New York.
Pommier T, Canback B, Riemann L, Bostrom KH, Simu K, Lundberg P et al. (2007). Global patterns of diversity and community structure in marine bacterioplankton. Mol Ecol 16: 867–880.
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1: 283–290.
Ronaghi M, Uhlen M, Nyren P . (1998). A sequencing method based on real-time pyrophosphate. Science 281: 363–365.
Rosson RA, Nealson KH . (1982). Manganese binding and oxidation by spores of a marine bacillus. J Bacteriol 151: 1027–1034.
Rothberg JM, Leamon JH . (2008). The development and impact of 454 sequencing. Nat Biotechnol 26: 1117–1124.
Schloss P, Handelsman J . (2005). Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71: 1501–1506.
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR et al. (2006). Microbial diversity in the deep sea and the underexplored rare biosphere. Proc Natl Acad Sci USA 103: 12115–12120.
Staley JT, Gosink JJ . (1999). Poles apart: biodiversity and biogeography of sea ice bacteria. Annu Rev Microbiol 53: 189–215.
Swallow J, Crease J . (1965). Hot salty water at the bottom of the Red Sea. Nature 205: 165–166.
Teske A, Sorensen KB . (2007). Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J 2: 3–18.
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE et al. (2008). A core gut microbiome in obese and lean twins. Nature, 480–484.
Ulloa O, Pantoja S . (2009). The oxygen minimum zone of the eastern South Pacific. Deep Sea Res Part II 56: 987–991.
Venter J, Remington K, Heidelberg J, Halpern A, Rusch D, Eisen J et al. (2004). Environmental genome shotgun sequencing of the Sargasso Sea. Science 304: 66–74.
Wang G, Kennedy SP, Fasiludeen S, Rensing C, DasSarma S . (2004). Arsenic resistance in Halobacterium sp. strain NRC-1 examined by using an improved gene knockout system. J Bacteriol 186: 3187–3194.
Wang Y, Qian P-Y . (2009). Conservative fragments in bacterial 16S rRNA genes and primer design for 16S ribosomal DNA amplicons in metagenomic studies. PLoS ONE 4: e7401.
Whitaker RJ, Grogan DW, Taylor JW . (2003). Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301: 976–978.
Wolfe R (2006). The Archaea: a personal overview of the formative years. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (Eds) The prokaryotes: a handbook on the biology of bacteria, vol. 3. Springer-Verlag: New York. pp. 3–9.
Zhou J, Bruns MA, Tiedje JM . (1996). DNA recovery from soils of diverse composition. Appl Environ Microbiol 62: 316–322.
Acknowledgements
We thank ML Sogin for his constructive comments on this article, the crew of the R/V Oceanus for providing technical assistance during the expedition and A Bower and S Swift of the Woods Hole Oceanographic Institution for providing environmental data and useful information of the study sites. We also thank R-L Liu of HKUST for the enumeration of microbial cells densities in water samples. This study was supported by an award (SA-C0040/UK-C0016) granted to PY Qian from the King Abdullah University of Science and Technology.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplementary Information accompanies the paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Qian, PY., Wang, Y., Lee, O. et al. Vertical stratification of microbial communities in the Red Sea revealed by 16S rDNA pyrosequencing. ISME J 5, 507–518 (2011). https://doi.org/10.1038/ismej.2010.112
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.112
Keywords
This article is cited by
-
Increased prokaryotic diversity in the Red Sea deep scattering layer
Environmental Microbiome (2023)
-
Water Mass Controlled Vertical Stratification of Bacterial and Archaeal Communities in the Western Arctic Ocean During Summer Sea-Ice Melting
Microbial Ecology (2023)
-
Assessing the Nutrient Dynamics in a Himalayan Warm Monomictic Lake
Water, Air, & Soil Pollution (2021)
-
Activity, Abundance, and Community Composition of Anaerobic Ammonia–Oxidizing (Anammox) Bacteria in Sediment Cores of the Pearl River Estuary
Estuaries and Coasts (2020)
-
Salinity stresses make a difference in the start-up of membrane bioreactor: performance, microbial community and membrane fouling
Bioprocess and Biosystems Engineering (2019)