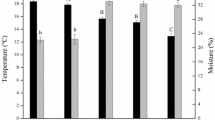
Abstract
Soils collected across a long-term liming experiment (pH 4.0–8.3), in which variation in factors other than pH have been minimized, were used to investigate the direct influence of pH on the abundance and composition of the two major soil microbial taxa, fungi and bacteria. We hypothesized that bacterial communities would be more strongly influenced by pH than fungal communities. To determine the relative abundance of bacteria and fungi, we used quantitative PCR (qPCR), and to analyze the composition and diversity of the bacterial and fungal communities, we used a bar-coded pyrosequencing technique. Both the relative abundance and diversity of bacteria were positively related to pH, the latter nearly doubling between pH 4 and 8. In contrast, the relative abundance of fungi was unaffected by pH and fungal diversity was only weakly related with pH. The composition of the bacterial communities was closely defined by soil pH; there was as much variability in bacterial community composition across the 180-m distance of this liming experiment as across soils collected from a wide range of biomes in North and South America, emphasizing the dominance of pH in structuring bacterial communities. The apparent direct influence of pH on bacterial community composition is probably due to the narrow pH ranges for optimal growth of bacteria. Fungal community composition was less strongly affected by pH, which is consistent with pure culture studies, demonstrating that fungi generally exhibit wider pH ranges for optimal growth.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Aciego Pietri JC, Brookes PC . (2007a). Nitrogen mineralisation along a pH gradient of a silty loam UK soil. Soil Biol Biochem 40: 797–802.
Aciego Pietri JC, Brookes PC . (2007b). Relationships between soil pH and microbial properties in a UK arable soil. Soil Biol Biochem 40: 1856–1861.
Aciego Pietri JC, Brookes PC . (2009). Substrate inputs and pH as factors controlling microbial biomass, activity and community structure in an arable soil. Soil Biol Biochem 41: 1396–1405.
Axelrood PE, Chow ML, Radomski CC, McDermott JM, Davies J . (2002). Molecular characterization of bacterial diversity from British Columbia forest soils subjected to disturbance. Can J Microbiol 48: 655–674.
Bååth E . (1996). Adaptation of soil bacterial communities to prevailing pH in different soils. FEMS Microb Ecol 19: 227–237.
Bååth E, Anderson TH . (2003). Comparison of soil fungal/bacterial ratios in a pH gradient using physiological and PLFA-based techniques. Soil Biol Biochem 35: 955–963.
Baker KL, Langenheder S, Nicol GW, Ricketts D, Killham K, Campbell CD et al. (2009). Environmental and spatial characterisation of bacterial community composition in soil to inform sampling strategies. Soil Biol Biochem 41: 2292–2298.
Beales N . (2004). Adaptation of microorganisms to cold temperature, weak acid preservatives, low pH, and osmotic stress: a review. Comp Rev Food Sci F 3: 1–20.
Bennett LT, Kasel S, Tibbits J . (2009). Woodland trees modulate soil resources and conserve fungal diversity in fragmented landscapes. Soil Biol Biochem 41: 2162–2169.
Borneman J, Hartin REJ . (2000). PCR primers that amplify fungal rRNA genes from environmental samples. Appl Environ Microbiol 66: 4356–4360.
Buée M, Reich M, Murat C, Morin E, Nilsson RH, Uroz S et al. (2009). 454 pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytol 184: 449–456.
Caporaso JS, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows integration and analysis of high-throughput community sequencing data. Nat Methods (in press), doi:10.1038/nmeth.f.303.
Davis JP, Youssef NH, Elshahed MS . (2009). Assessment of the diversity, abundance, and ecological distribution of candidate division SR1 reveals a high level of phylogenetic diversity but limited morphotypic diversity. Appl Environ Microbiol 75: 4139–4148.
Dimitriu PA, Grayston SJ . (2010). Relationship between soil properties and patterns of bacterial β-diversity across reclaimed and natural boreal forest soils. Microb Ecol 59: 563–573.
Domsch KH, Gams W, Anderson TH . (1980). Compendium of Soil Fungi Volume 1 Academic Press: London.
Fazi S, Amalfitano S, Pernthaler J, Puddu A . (2005). Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environ Microbiol 7: 1633–1640.
Fernández-Calviño D, Bååth E . (2010). Growth response of the bacterial community to pH in soils differing in pH. FEMS Microbiol Ecol (in press), doi: 10.1111/j.1574-6941.2010.00873.x.
Fierer N, Bradford MA, Jackson RB . (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364.
Fierer N, Hamady M, Lauber CL, Jackson RB . (2008). The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci USA 105: 17994–17999.
Fierer N, Jackson RB . (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631.
Fierer N, Jackson JA, Vilgalys R, Jackson RB . (2005). Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71: 4117–4120.
Fierer N, Strickland MS, Lipzin D, Bradford MA, Cleveland CC . (2009). Global patterns in belowground communities. Ecol Lett 12: 1238–1249.
Frey SD, Knorr M, Parrent JL, Simpson RT . (2004). Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. For Ecol Manag 196: 159–171.
Hamady M, Lozupone C, Knight R . (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27.
Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R . (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237.
Hartman WH, Richardson CJ, Vilgalys R, Bruland GL . (2008). Environmental and anthropogenic control of bacterial communities in wetland soils. Proc Natl Acad Sci USA 105: 17842–17847.
Jenkins SN, Waite IS, Blackburn A, Husband R, Rushton SP, Manning DC et al. (2009). Actinobacterial community dynamics in long term managed grasslands. 95: 319–334.
Jesus ED, Marsh TL, Tiedje JM, Moreira FMD . (2009). Changes in lands use alter the structure of bacterial communities in Western Amazon soils. ISME J 3: 1004–1011.
Jones RT, Robeson MS, Lauber CL, Hamady M, Knight R, Fierer N . (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J 3: 442–453.
Kirchman DL . (2002). The ecology of Cytophaga–Flavobacteria in aquatic environments. FEMS Microbiol Ecol 39: 91–100.
Kunin V, Engelbrektson A, Ochman H, Hugenholtz P . (2010). Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12: 118–123.
Lauber CL, Hamady M, Knight R, Fierer N . (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community composition at the continental scale. Appl Environ Microbiol 75: 5111–5120.
Lauber CL, Strickland MS, Bradford MA, Fierer N . (2008). The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40: 2407–2415.
Li W, Godzik A . (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659.
Lozupone C, Hamady M, Knight R . (2006). UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 731.
Lozupone C, Knight R . (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235.
Männistö MK, Tiirola M, Häggblom MM . (2007). Bacterial communities in Arctic fjelds of Finnish Lapland are stable but highly pH dependent. FEMS Microbiol Ecol 59: 452–465.
McCaig AE, Glover LA, Prosser JI . (1999). Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol 65: 1721–1730.
Nevarez L, Vasseur V, Le Madec L, Le Bras L, Coroller L, Leguérinel I et al. (2009). Physiological traits of Penicillium glabrum strain LCP 08.5568, a filamentous fungus isolated from bottled aromatised mineral water. Int J Food Microbiol 130: 166–171.
Nicol GW, Leininger S, Schleper C, Prosser JI . (2008). The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ Microbiol 10: 2966–2978.
Nilsson LO, Bååth E, Falkengren-Grerup U, Wallander H . (2007). Growth of ectomycorrhizal mycelia and composition of soil microbial communities in oak forest soils along a nitrogen deposition gradient. Oecologia 153: 375–384.
Philippot L, Cuhel J, Saby NPA, Chèneby D, Chronáková A, Bru D et al. (2009). Mapping fine-scale spatial patterns of size and activity of the denitrifier community. Environ Microbiol 11: 1518–1526.
Price MN, Dehal PS, Arkin AP . (2009). Fasttree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650.
Pruesse E, Quast C, Knittel K, Fuchs B, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196.
Rosso L, Lobry JR, Bajard S, Flandrois JP . (1995). Convenient model to describe the combined effects of temperature and pH on microbial growth. Appl Environ Microbiol 61: 610–616.
Rousk J, Aldén Demoling L, Bahr A, Bååth E . (2008). Examining the fungal and bacterial niche overlap using selective inhibitors in soil. FEMS Microbiol Ecol 63: 350–358.
Rousk J, Brookes PC, Bååth E . (2009). Contrasting soil pH effects on fungal and bacterial growth suggests functional redundancy in carbon mineralisation. Appl Environ Microbiol 75: 1589–1596.
Rousk J, Brookes PC, Bååth E . (2010a). The microbial PLFA composition as affected by pH in an arable soil. Soil Biol Biochem 42: 516–520.
Rousk J, Brookes PC, Bååth E . (2010b). Investigating the mechanisms for the opposing pH-relationships of fungal and bacterial growth in soil. Soil Biol Biochem 42: 926–934.
Shaw AK, Halpern AL, Beeson K, Tran B, Venter JC, Martiny JBH . (2008). It's all relative: ranking the diversity of aquatic bacterial communities. Environ Microbiol 10: 2200–2210.
USDA (United States Department of Agriculture Soil Survey Staff (1992). Keys to soil taxonomy. SMSS Technical Monograph 19. Pocahontas Press: Blacksburg, VA, 541.
Wheeler KA, Hurdman BF, Pitt JI . (1991). Influence of pH on the growth of some toxigenic species of Aspergillus, Penicillium and Fusarium. Int J Food Microbiol 12: 141–150.
Acknowledgements
We thank D Ahrén for helpful discussions and M Hamady for assistance with bioinformatics. This study was supported by grants from the Swedish Research Council (VR) and from the Royal Physiographic Society of Lund (Kgl Fysiografen) (to JR). This study was also supported by grants from the US National Science Foundation, the US Department of Agriculture and the AW Mellon Foundation to NF and by a grant from the Swedish Research Council (VR) to EB
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rousk, J., Bååth, E., Brookes, P. et al. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J 4, 1340–1351 (2010). https://doi.org/10.1038/ismej.2010.58
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2010.58
Keywords
This article is cited by
-
Effects of simulated atmospheric nitrogen deposition on the bacterial community structure and diversity of four distinct biocolonization types on stone monuments: a case study of the Leshan Giant Buddha, a world heritage site
Heritage Science (2024)
-
Land management shapes drought responses of dominant soil microbial taxa across grasslands
Nature Communications (2024)
-
Pervasive associations between dark septate endophytic fungi with tree root and soil microbiomes across Europe
Nature Communications (2024)
-
Vertical differences in carbon metabolic diversity and dominant flora of soil bacterial communities in farmlands
Scientific Reports (2024)
-
Identifying key soil characteristics for Francisella tularensis classification with optimized Machine learning models
Scientific Reports (2024)