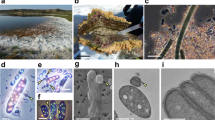
Abstract
Vampire amoebae (vampyrellids) are predators of algae, fungi, protozoa and small metazoans known primarily from soils and in freshwater habitats. They are among the very few heterotrophic naked, filose and reticulose protists that have received some attention from a morphological and ecological point of view over the last few decades, because of the peculiar mode of feeding of known species. Yet, the true extent of their biodiversity remains largely unknown. Here we use a complementary approach of culturing and sequence database mining to address this issue, focusing our efforts on marine environments, where vampyrellids are very poorly known. We present 10 new vampyrellid isolates, 8 from marine or brackish sediments, and 2 from soil or freshwater sediment. Two of the former correspond to the genera Thalassomyxa Grell and Penardia Cash for which sequence data were previously unavailable. Small-subunit ribosomal DNA analysis confirms they are all related to previously sequenced vampyrellids. An exhaustive screening of the NCBI GenBank database and of 454 sequence data generated by the European BioMarKs consortium revealed hundreds of distinct environmental vampyrellid sequences. We show that vampyrellids are much more diverse than previously thought, especially in marine habitats. Our new isolates, which cover almost the full phylogenetic range of vampyrellid sequences revealed in this study, offer a rare opportunity to integrate data from environmental DNA surveys with phenotypic information. However, the very large genetic diversity we highlight within vampyrellids (especially in marine sediments and soils) contrasts with the paradoxically low morphological distinctiveness we observed across our isolates.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ . (1990). Basic local alignment search tool. J Mol Biol 215: 403–410.
Bass D, Cavalier-Smith T . (2004). Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa). Int J Syst Evol Microbiol 54: 2393–2404.
Bass D, Chao EEY, Nikolaev S, Yabuki A, Ishida K, Berney C et al (2009). Phylogeny of novel naked filose and reticulose Cercozoa: Granofilosea cl. n. and Proteomyxidea revised. Protist 160: 75–109.
Bass D, Yabuki A, Santini S, Romac S, Berney C . (2012). Reticulamoeba is a long-branched Granofilosean (Cercozoa) that is missing from sequence databases. PLoS One 7: e49090.
Berney C, Fahrni J, Pawlowski J . (2004). How many novel eukaryotic ‘kingdoms’? Pitfalls and limitations of environmental DNA surveys. BMC Biol 2: 13.
Boenigk J, Ereshefsky M, Hoef-Emden K, Mallet J, Bass D . (2012). Concepts in protistology: species definitions and boundaries. Eur J Protistol 48: 96–102.
Bonkowski M, Cheng W, Griffiths BS, Alphei J, Scheu S . (2000). Microbial-faunal interactions in the rhizosphere and effects on plant growth. Eur J Soil Biol 36: 135–147.
Cash J . (1904). On some new and little-known British freshwater Rhizopoda. J Linn Soc (Zool) 24: 218–225.
Cavalier-Smith T . (1998). A revised six-kingdom system of life. Biol Rev Camb Philos Soc 73: 203–266.
Cavalier-Smith T . (2002). The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol 52: 297–354.
Cienkowski L . (1865). Beiträge zur Kenntnis der Monaden. Arch Mikrosk Anat 1: 203–232.
Cienkowski L . (1876). Über einige Rhizopoden und verwandte Organismen. Arch Mikrosk Anat 12: 15–50.
Dawson SC, Pace NR . (2002). Novel kingdom-level eukaryotic diversity in anoxic environments. Proc Natl Acad Sci USA 99: 8324–8329.
Dobell C . (1913). Observations on the life-history of Cienkowski's “Arachnula”. Arch Protistenkd 31: 317–353.
Felsenstein J . (1981). Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17: 368–376.
Felsenstein J . (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Grell KG . (1985). Der Formwechsel des plasmodialen Rhizopoden Thalassomyxa australis n.g., n.sp. Protistologica 21: 215–233.
Grell KG . (1992). A species of Thalassomyxa from the North coast of Jamaica. Arch Protistenkd 142: 15–33.
Hall TA . (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98.
Hertwig R, Lesser E . (1874). Über Rhizopoden und denselben nahestchenden Organismen. Arch Mikrosk Anat 10: (Suppl) 61–65.
Hess S, Sausen N, Melkonian M . (2012). Shedding light on vampires: the phylogeny of vampyrellid amoebae revisited. PLoS One 7: e31165.
Huelsenbeck JP, Ronquist F . (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755.
Hülsmann N . (1993). Lateromyxa gallica n. g., n. sp. (Vampyrellidae): a filopodial amoeboid protist with a novel life cycle and conspicuous ultrastructural characters. J Euk Microbiol 40: 141–149.
Lanave C, Preparata G, Saccone C, Serio G . (1984). A new method for calculating evolutionary substitution rates. J Mol Evol 20: 86–93.
Lara E, Mitchell EAD, Moreira D, López García P . (2011). Highly diverse and seasonally dynamic protest community in a pristine peat bog. Protist 162: 14–32.
Lecroq B, Gooday AJ, Cedhagen T, Sabbatini A, Pawlowski J . (2009). Molecular analyses reveal high levels of eukaryotic richness associated with enigmatic deep-sea protists (Komokiacea). Mar Biodiv 39: 45–55.
Lloyd FE . (1929). The behavior of Vampyrella lateritia, with special reference to the work of Professor Chr. Gobi. Arch Protistenkd 67: 219–236.
Logares R, Audic S, Santini S, Pernice MC, de Vargas C, Massana R . (2012). Diversity patterns and activity of uncultured marine heterotrophic flagellates unveiled with pyrosequencing. ISME J 6: 1823–1833.
Logares R, Bråte J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K . (2009). Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol 17: 414–422.
Old KM . (1978). Perforation and lysis of fungal spores by soil amoebae. Ann Appl Biol 89: 128–131.
Old KM, Chakraborty S . (1986). Mycophagous soil amoebae: their biology and significance in the ecology of soil-borne plant pathogens. Prog Protistol 1: 163–194.
Old KM, Darbyshire JF . (1980). Arachnula impatiens Cienk., a mycophagous giant amoeba from soil. Protistologica 16: 277–287.
Patterson DJ . (1999). The diversity of Eukaryotes. Am Nat 154: S96–S124.
Patterson DJ, Simpson AGB, Rogerson A . (2000). Amoebae of uncertain affinities. In: Lee JJ, Leedale GF, Bradbury P, (eds) The Illustrated Guide to the Protozoa 2nd edn Society of Protozoologists: Lawrence, KS, USA, pp 804–827.
Rodriguez F, Oliver JL, Marin A, Medina JR . (1990). The general stochastic model of nucleotide substitution. J Theor Biol 142: 485–501.
Rodríquez-Martínez R, Rocap G, Logares R, Romac S, Massana R . (2012). Low evolutionary diversification in a widespread and abundant uncultured protist (MAST-4). Mol Biol Evol 29: 1393–1406.
Ronquist F, Huelsenbeck JP . (2003). MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Röpstorf P, Hülsmann N, Hausmann K . (1994). Comparative fine structural investigations of interphase and mitotic nuclei of vampyrellid filose amoebae. J Euk Microbiol 41: 18–30.
Schepotieff A . (1912). Untersuchungen über niedere Organismen. III. Monerenstudien. Zool Jahrb (Abt f Anat) 32: 367–400.
Stamatakis A . (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690.
Tkach V, Pawlowski J . (1999). A new method of DNA extraction from the ethanol-fixed parasitic worms. Acta Parasitol 44: 147–148.
Wegener Parfrey L, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML et al (2010). Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol 59: 518–533.
Wuyts J, De Rijk P, Van de Peer Y, Pison G, Rousseeuw P, De Wachter R . (2000). Comparative analysis of more than 3000 sequences reveals the existence of two pseudoknots in area V4 of eukaryotic small subunit ribosomal RNA. Nucleic Acids Res 28: 4698–4708.
Zopf W . (1885). Die Pilzthiere oder Schleimpilze. In: Schenk A, (ed) Handbuch des Botanik, Enzyklopädie der Naturwissenschaften Vol. 3. Eduard Trewendt: Breslau, pp 1–174.
Zwillenberg LO . (1953). Theratromyxa weberi, a new proteomyxean organism from soil. Ant Leeuw (J Microbiol Serol) 19: 101–116.
Acknowledgements
We thank NERC for a Standard Research Grant (NE/H009426/1) (DB and CB) and a New Investigator Grant (NE/H000887/1) (DB). We are grateful to Tom Cavalier-Smith, Ema Chao, Libby Snell, Joe Carlson and Josephine Scoble for help with sampling. SS is grateful to Hiroyuki Ogata for support and advice about phylogenetic mapping, and Professor Jean-Michel Claverie for providing access to the computer facility of the PACA-Bioinfo IBISA platform. The IGS laboratory is partially supported by the French ANR grant ANT-08-BDVA-003. The BioMarKs 454 sequence data were generated as part of a study supported by the EU-FP7 ERA-net program BiodivERsA, under the project BioMarKs (2008-6530). We thank all additional members of the BioMarKs consortium who participated to field sampling and data generation (in particular Stéphane Audic and the coordinator of the project Colomban de Vargas), as well as three anonymous reviewers for very useful comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on The ISME Journal website
Supplementary information
Rights and permissions
About this article
Cite this article
Berney, C., Romac, S., Mahé, F. et al. Vampires in the oceans: predatory cercozoan amoebae in marine habitats. ISME J 7, 2387–2399 (2013). https://doi.org/10.1038/ismej.2013.116
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ismej.2013.116
Keywords
This article is cited by
-
The impact of protozoa addition on the survivability of Bacillus inoculants and soil microbiome dynamics
ISME Communications (2022)
-
Various brain-eating amoebae: the protozoa, the pathogenesis, and the disease
Frontiers of Medicine (2021)
-
Marinomyxa Gen. Nov. Accommodates Gall-Forming Parasites of the Tropical to Subtropical Seagrass Genus Halophila and Constitutes a Novel Deep-Branching Lineage Within Phytomyxea (Rhizaria: Endomyxa)
Microbial Ecology (2021)
-
First report of vampyrellid predator–prey dynamics in a marine system
The ISME Journal (2019)
-
The disappearing periglacial ecosystem atop Mt. Kilimanjaro supports both cosmopolitan and endemic microbial communities
Scientific Reports (2019)