Abstract
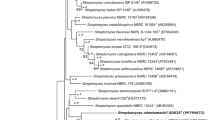
An actinomycete strain, IR73-Li102T, was isolated from a lichen sample obtained from Iriomote Island, Japan, and subsequently characterized using a polyphasic approach. Comparative 16S rRNA gene sequence analysis revealed that strain IR73-Li102T had the highest sequence similarities with Actinomycetospora chiangmaiensis YIM 0006T (98.3%), A. corticola 014-5T (98.1%) and A. rishiriensis RI109-Li102T (98.0%). However, DNA–DNA hybridization assays, as well as physiological and biochemical analyzes, showed that strain IR73-Li102T could be clearly differentiated from its closest phylogenetic relatives. The strain contained meso-diaminopimelic acid, and arabinose and galactose were present in whole-cell hydrolysates. The predominant menaquinone was MK-8(H4), and the diagnostic phospholipids were phosphatidylethanolamine, phosphatidylcholine, phosphatidylinositol and diphosphatidylglycerol. The major cellular fatty acid was iso-C16:0 (58%). The chemotaxonomic properties of strain IR73-Li102T were consistent with those shared by members of the genus Actinomycetospora. On the basis of the phenotypic features and DNA–DNA hybridization data, strain IR73-Li102T (= NBRC 106365T = KCTC 19783T) represents a novel species of the genus Actinomycetospora, for which the name Actinomycetospora iriomotensis sp. nov. is proposed.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
Accession codes
References
Mandrioli, P., Caneva, G. & Sabbioni, C. Cultural Heritage and Aerobiology: Methods and Measurement Techniques for Biodeterioration Monitoring (Springer, Verlag Berlin Heidelberg, 2003).
Nash, T. H. Lichen Biology (Cambridge University Press, Cambridge, UK, 1996).
González, I., Ayuso-Sacido, A., Anderson, A. & Genilloud, O. Actinomycetes isolated from lichens: evaluation of their diversity and detection of biosynthetic gene sequences. FEMS Microbiol. Ecol. 54, 401–415 (2005).
Cardinale, M., Puglia, A. M. & Grube, M. Molecular analysis of lichen-associated bacterial communities. FEMS Microbiol. Ecol. 57, 484–495 (2006).
Motohashi, K., Takagi, M., Yamamura, H., Hayakawa, M. & Shin-ya, K. A new angucycline and a new butenolide isolated from lichen-derived Streptomyces spp. J. Antibiot. 63, 545–548 (2010).
Li, B., Xie, C.- H. & Yokota, A. Nocardioides exalbidus sp. nov., a novel actinomycete isolated from lichen in Izu-Oshima Island, Japan. Actinomycetologica 21, 22–26 (2007).
An, S.- Y., Xiao, T. & Yokota, A. Leifsonia lichenia sp. nov., isolated from lichen in Japan. J. Gen. Appl. Microbiol. 55, 339–343 (2009).
Yamamura, H. et al. Actinomycetospora rishiriensis sp. nov., an actinomycete isolated from a lichen. Int. J. Syst. Evol. Microbiol. (e-pub ahead of print 10 December 2010; doi:10.1099/ijs.0.028753-0).
Jiang, Y. et al. Actinomycetospora chiangmaiensis gen. nov., sp. nov., a new member of the family Pseudonocardiaceae. Int. J. Syst. Evol. Microbiol. 58, 408–413 (2008).
Tamura, T. et al. Actinomycetospora chibensis sp. nov., Actinomycetospora chloros sp. nov., Actinomycetospora cinnamomea sp. nov., Actinomycetospora corticola sp. nov., Actinomycetospora lutea sp. nov., Actinomycetospora straminea sp. nov. and Actinomycetospora succinea sp. nov. and emendation of the genus Actinomycetospora. Int. J. Syst. Evol. Microbiol. (e-pub ahead of print 9 July 2010; doi:10.1099/ijs.0.018366-0).
Hayakawa, M. & Nonomura, H. Humic acid-vitamin agar, a new medium for the selective isolation of soil actinomycetes. J. Ferment. Technol. 65, 501–509 (1987).
Hayakawa, M., Iino, S. & Nonomura, H. Heavy metal resistance and melanoid pigment production in the streptomycete flora of copper-polluted vineyard soils. Hakkokogaku 60, 1–9 (1982).
Gerhardt, P. Manual of Methods for General Bacteriology (American Society for Microbiology, Washington, DC, 1981).
Shirling, E. B. & Gottlieb, D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Williams, S. T. et al. Numerical classication of Streptomyces and related taxa. J. Gen. Microbiol. 129, 1743–1813 (1983).
Gordon, R. E. & Mihm, J. M. Identification of Nocardia caviae (Erikson) nov. comb. Ann. NY. Acad. Sci. 98, 628–636 (1962).
Hasegawa, T., Takizawa, M. & Tanida, S. A rapid analysis for chemical grouping of aerobic actinomycetes. J. Gen. Appl. Microbiol. 29, 319–322 (1983).
Schaal, K. P. Chemical Methods in Bacterial Systematics (Academic Press, London, 1985).
Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids. Microbial ID, Inc. (Newark, Delaware, 1990).
Minnikin, D. E. et al. An integrated procedure for the extraction of bacterial isoprenoid quinines and polar lipids. J. Microbiol. Methods 2, 233–241 (1984).
Tamaoka, J. & Komagata, K. Determination of DNA base composition by reversed- phase high-performance liquid chromatography. FEMS Microbiol. Lett. 25, 125–128 (1984).
Saito, H. & Miura, K.- I. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta. 72, 619–629 (1963).
Hatano, K., Nishii, T. & Kasai, H. Taxonomic re-evaluation of whorl-forming Streptomyces (formerly Streptoverticillium) species by using phenotype, DNA–DNA hybridization and sequences of gyrB, and proposal of Streptomyces luteireticuli (ex Katoh and Arai 1957) corring., sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 53, 1519–1529 (2003).
Tamura, T. & Hatano, K. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2119–2125 (2001).
Chun, J. et al. EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261 (2007).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Takahashi, K. & Nei, M. Efficiencies of fast algorithms of phylogenetic inference under the criteria of maximum parsimony, minimum evolution, and maximum likelihood when a large number of sequences are used. Mol. Biol. Evol. 17, 1251–1258 (2000).
Guindon, S., Lethiec, F., Duroux, P. & Gascuel, O. PHYML Online–a web server for fast maximum likelihood-based phylogenetic inference (Web Server issue). Nucleic Acids Res. 33, W557–W559 (2005).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 (1985).
Kusunoki, S. et al. Application of calorimetric microdilution plate hybridization for rapid generic identification of 22 Mycobacterium species. J. Clin. Microbiol. 29, 1596–1603 (1991).
Lechevalier, M. P. & Lechevalier, H. A. Chemical composition as a criterion in the classification of aerobic actinomycetes. Int. J. Syst. Evol. Microbiol. 20, 435–443 (1970).
Lechevalier, M. P., De Bivre, C. & Lechevalier, H. A. Chemotaxonomy of aerobic actinomycetes: phospholipid composition. Biochem. Syst. Ecol. 5, 249–260 (1977).
Wayne, L. G. et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464 (1987).
Acknowledgements
We are grateful to Mr Yuya Sakuraki for technical assistance. This study was supported in part by a research grant from the Institute for Fermentation, Osaka (IFO), Japan. We are grateful to Dr Jean P Euzéby for support with nomenclature.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yamamura, H., Ashizawa, H., Nakagawa, Y. et al. Actinomycetospora iriomotensis sp. nov., a novel actinomycete isolated from a lichen sample. J Antibiot 64, 289–292 (2011). https://doi.org/10.1038/ja.2011.15
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/ja.2011.15
Keywords
This article is cited by
-
Actinomycetospora termitidis sp. nov., an insect-derived actinomycete isolated from termite (Odontotermes formosanus)
The Journal of Antibiotics (2024)
-
Actinomycetospora callitridis sp. nov., an endophytic actinobacterium isolated from the surface-sterilised root of an Australian native pine tree
Antonie van Leeuwenhoek (2019)
-
Diversity, Antimicrobial Activity, and Biosynthetic Potential of Cultivable Actinomycetes Associated with Lichen Symbiosis
Microbial Ecology (2017)
-
Lichens as natural sources of biotechnologically relevant bacteria
Applied Microbiology and Biotechnology (2016)
-
Nutrient scavenging activity and antagonistic factors of non-photobiont lichen-associated bacteria: a review
World Journal of Microbiology and Biotechnology (2016)