Summary
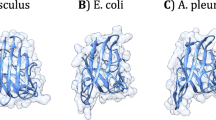
Chymotrypsinogen is widely present in various animal pancreases. To study evolutionary relationship of chymotrypsinogen gene in species, we used a cDNA probe of human prechymotrypsinogen to investigate the species distribution of chymotrypsinogen gene, and designed oligodeoxynucleotide primers to investigate the genomic organization in the three domains of active sites. The genomic analyses showed that chymotrypsinogen gene is evolutionary conserved in species. On the basis of the deduced amino acid residues, a three-dimensional model for human chymotrypsinogen was further built by computer graphics. The model showed high similarity to the X-ray crystal structure of bovine chymotrypsinogen A, thus, demonstrated that the three-dimensional structure is more conserved in evolution than protein sequences.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Bell GI, Quinto C, Quiroga M, Valenzuela P, Craik CS, Rutter W (1984): Isolation and sequence of a rat chymotrypsin B gene. J Biol Chem259: 14265–14270
Bhargava AK, Barnard EA (1973): Evolution in the pancreatic chymotrypsinogen series: N-terminal sequence determinations and comparisons. J Mol Biol2: 187–198
Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M (1977): The Protein Data Bank: A computer-based archival file for macromolecular structures. J Mol Biol112: 535–542
Caro AD, Figarella, C, Guy O (1975): The two human chymotrypsinogens purification and characterization. Biochim Biophys Acta379: 431–443
Chothia C, Lesk AM (1986): The relation between the divergence of sequence and structure in protein. EMBO J5: 823–826
Cohen GH, Silverton EW, Davier DR (1981): Refined crystal structure of gamma-chymotrypsin at 1.9 angstroms resolution. J Mol Biol148: 449–479
Feinberg AP, Vogelstein B (1983): A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem132: 6–13
Haneef I, Talbot SJ, Stockley PG (1989): Modeling loop structures in proteins and nucleic acids: an RNA stem-loop. J Mol Graphics7: 186–195
Haën CD, Neurath H, Teller DC (1975): The phylogeny of trypsin-related serine protease and their zymogens. New methods for the investigation of distant evolutionary relationship. J Mol Biol92: 225–259
Hou D-X, Ozawa K, Tomita N, Maeda Y, Hashiguchi T, Yokoyama K, Soeda E (1993): Genomic cloning and partial characterization of human chymotrypsinogen gene. Jpn J Human Genet38: 371–380
Maniatis T, Fritsch EF, Sambrook J (1989): Molecular Cloning: A Laboratory Manual (2nd ed), Cold Spring Harbor Laboratory Press, New York
Neurath H, Walsh KA, Winter WP (1967): Evolution of structure and function of proteinase. Science158: 1638–1644
Pinsky SD, LaForge S, Luc V, Scheele G (1993): Identification of cDNA clones encoding secretory isoenzyme forms: Sequence determination of canine pancreatic prechymotrypsinogen 2 mRNA. Proc Natl Acad Sci USA80: 7486–7490
Ryan CA, John JC, Tomimatsu Y (1965): Chicken chymotrypsin and turkey trypsin, Part II: physical and enzyme properties. Arch Biochem Biophys110: 175–183
Tomita N, Izumoto Y, Horii A, Doi S, Yokouchi H, Ogawa M, Mori T, Matsubara, K (1989): Molecular cloning and nucleotide sequence of human pancreatic prechymotrypsinogen cDNA. Biochem Biophys Res Commun158: 569–574
Wang D, Bode W, Huber R (1985): Bovine chymotrypsinogen A X-ray crystal structure analysis and refinement of a new crystal form at 1.8 Å resolution. J Mol Biol158: 595–624
Wilcox PE (1970): Chymotrypsinogens-chymotrypsin. Perlmann GE, Lorand L (eds). Methods in enzymology, Vol 19, pp 64–108, Academic Press, New York
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hou, DX., Wang, Y., Yamashita, H. et al. Evolutionary conservation of chymotrypsinogen gene: Genomic analysis and protein modeling. Jap J Human Genet 39, 235–242 (1994). https://doi.org/10.1007/BF01876843
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1007/BF01876843