Abstract
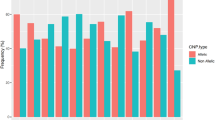
DNA-repair pathways are critical for maintaining the integrity of the genetic material by protecting against mutations due to exposure-induced damages or replication errors. Polymorphisms in the corresponding genes may be relevant in genetic epidemiology by modifying individual cancer susceptibility or therapeutic response. We report data on the population distribution of potentially functional variants in XRCC1, APEX1, ERCC2, ERCC4, hMLH1, and hMSH3 genes among groups representing individuals of European, Middle Eastern, African, Southeast Asian and North American descent. The data indicate little interpopulation differentiation in some of these polymorphisms and typical F ST values ranging from 10 to 17% at others. Low F ST was observed in APEX1 and hMSH3 exon 23 in spite of their relatively high minor allele frequencies, which could suggest the effect of balancing selection. In XRCC1, hMSH3 exon 21 and hMLH1 Africa clusters either with Middle East and Europe or with Southeast Asia, which could be related to the demographic history of human populations, whereby human migrations and genetic drift rather than selection would account for the observed differences.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Abdel-Rahman SZ, Soliman AS, Bondy ML, Omar S, El-Badawy SA, Khaled HM, Seifeldin IA, Levin B (2000) Inheritance of the 194Trp and the 399Gln variant alleles of the DNA repair gene XRCC1 are associated with increased risk of early-onset colorectal carcinoma in Egypt. Cancer Lett 159:79–86
Benachenhou N, Guiral S, Gorska-Flipot I, Labuda D, Sinnett D (1998) High resolution deletion mapping reveals frequent allelic losses at the DNA mismatch repair loci hMLH1 and hMSH3 in non-small cell lung cancer. Int J Cancer 77:173–180
Bessho T, Sancar A, Thompson LH, Thelen MP (1997) Reconstitution of human excision nuclease with recombinant XPF-ERCC1 complex. J Biol Chem 272:3833–3837
Bürger R (2000) The mathematical theory of selection, recombination, and mutation, Wiley, New York
Butkiewicz D, Rusin M, Enewold L, Shields PG, Chorazy M, Harris CC (2001) Genetic polymorphisms in DNA repair genes and risk of lung cancer. Carcinogenesis 22:593–597
Caldecott KW, Aoufouchi S, Johnson P, Shall S (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res 24:4387–4394
Cavalli-Sforza LL (1966) Population structure and human evolution. Proc R Soc Lond B Biol Sci 164:362–379
Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes, Princeton University Press, Princeton
Crompton NE, Ozsahin M (1997) A versatile and rapid assay of radiosensitivity of peripheral blood leukocytes based on DNA and surface-marker assessment of cytotoxicity. Radiat Res 147:55–60
Demple B, Herman T, Chen DS (1991) Cloning and expression of APE, the cDNA encoding the major human apurinic endonuclease: definition of a family of DNA repair enzymes. Proc Natl Acad Sci U S A 88:11450–11454
Duell EJ, Millikan RC, Pittman GS, Winkel S, Lunn RM, Tse CK, Eaton A, Mohrenweiser HW, Newman B, Bell DA (2001) Polymorphisms in the DNA repair gene XRCC1 and breast cancer. Cancer Epidemiol Biomarkers Prev 10:217–222
Dybdahl M, Vogel U, Frentz G, Wallin H, Nexo BA (1999) Polymorphisms in the DNA repair gene XPD: correlations with risk and age at onset of basal cell carcinoma. Cancer Epidemiol Biomarkers Prev 8:77–81
Fan F, Liu C, Tavare S, Arnheim N (1999) Polymorphisms in the human DNA repair gene XPF. Mutat Res 406:115–120
Fredman D, Siegfried M, Yuan YP, Bork P, Lehvaslaiho H, Brookes AJ (2002) HGVbase: a human sequence variation database emphasizing data quality and a broad spectrum of data sources. Nucleic Acids Res 30:387–391
Fullerton SM, Bartoszewicz A, Ybazeta G, Horikawa Y, Bell GI, Kidd KK, Cox NJ, Hudson RR, Di Rienzo A (2002) Geographic and haplotype structure of candidate type 2 diabetes susceptibility variants at the calpain-10 locus. Am J Hum Genet 70:1096–1106
Hartl DL, Clark AG (1989) Principles of population genetics, Sinauer, Sunderland
Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir MR, Ferak V, Villems R (1999) Deep common ancestry of indian and western-Eurasian mitochondrial DNA lineages. Curr Biol 9:1331–1334
Kolodner R (1996) Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev 10:1433–1442
Labuda D, Krajinovic M, Richer C, Skoll A, Sinnett H, Yotova V, Sinnett D (1999) Rapid detection of CYP1A1, CYP2D6, and NAT variants by multiplex polymerase chain reaction and allele-specific oligonucleotide assay. Anal Biochem 275:84–92
Lahr MM, Foley RA (1994) Multiple dispersals and modern human origins. Evol Anthropol 3:48–60
Lehmann AR (1995) Nucleotide excision repair and the link with transcription. Trends Biochem Sci 20:402–405
Lewis PO, Zaykin D (2001) Genetic data analysis: Computer program for the analysis of allelic data. v. 1.0 (d16c)
Lewontin RC, Krakauer J (1973) Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74:175–195
Liu B, Nicolaides NC, Markowitz S, Willson JK, Parsons RE, Jen J, Papadopolous N, Peltomaki P, de la Chapelle A, Hamilton SR, et al (1995) Mismatch repair gene defects in sporadic colorectal cancers with microsatellite instability. Nat Genet 9:48–55
Lunn RM, Langlois RG, Hsieh LL, Thompson CL, Bell DA (1999) XRCC1 polymorphisms: effects on aflatoxin B1-DNA adducts and glycophorin A variant frequency. Cancer Res 59:2557–2561
Mohrenweiser HW, Jones IM (1998) Variation in DNA repair is a factor in cancer susceptibility: a paradigm for the promises and perils of individual and population risk estimation? Mutat Res 400:15–24
Neel JV (1962) Diabetes mellitus: a “thrifty” genotype rendered detrimental by “progress”? Am J Hum Genet 14:353–362
Pero RW, Bryngelsson C, Bryngelsson T, Norden A (1983) A genetic component of the variance of N-acetoxy-2-acetylaminofluorene-induced DNA damage in mononuclear leukocytes determined by a twin study. Hum Genet 65:181–184
Pero RW, Johnson DB, Markowitz M, Doyle G, Lund-Pero M, Seidegard J, Halper M, Miller DG (1989) DNA repair synthesis in individuals with and without a family history of cancer. Carcinogenesis 10:693–697
Quintana-Murci L, Semino O, Bandelt HJ, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441
Relethford JH (2001) Genetics and the search for modern human origins, Wiley, New York
Rosenberg N, Murata M, Ikeda Y, Opare-Sem O, Zivelin A, Geffen E, Seligsohn U (2002) The frequent 5,10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in whites, Japanese, and Africans. Am J Hum Genet 70:758–762
Schneider S, Roessli D, Excoffier L (2000) Arlequin: A software for population genetics data analysis. v. 2.000. Genetics and Biometry Laboratory, Department of Anthropology, University of Geneva, Switzerland
Shen MR, Jones IM, Mohrenweiser H (1998) Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res 58:604–608
Sung P, Bailly V, Weber C, Thompson LH, Prakash L, Prakash S (1993) Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature 365:852–855
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgements
We are grateful to our colleagues who shared samples from their collections and to all individuals who kindly consented to provide DNA for this study. We thank Chantal Richer, Hugues Sinnett, Patrick Beaulieu, and Vania Yotova for their assistance, and Dominika Kozubska for secretarial help. MK and DS are scholars of the Fonds de la Recherche en Santé du Québec, whereas GM and CM were recipients of studentships from the Fondation Hôpital Ste-Justine/Power Corporation Inc., and the Natural Sciences and Engineering Research Council, respectively. This work was supported by the Fondation Charles-Bruneau and, in part, by a research grant from the Canadian Institutes of Health Research to DL (MOP-12782).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mathonnet, G., Labuda, D., Meloche, C. et al. Variable continental distribution of polymorphisms in the coding regions of DNA-repair genes. J Hum Genet 48, 659–664 (2003). https://doi.org/10.1007/s10038-003-0097-0
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1007/s10038-003-0097-0