Abstract
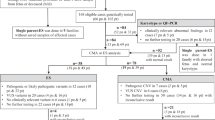
Cell-free fetal DNA (cffDNA) in the supernatant of amniotic fluid, which is usually discarded, can be used as a sample for prenatal diagnosis. For rapid prenatal diagnosis of frequent chromosome abnormalities, for example trisomies 13, 18, and 21, and monosomy X, using cffDNA, we have developed a targeted microarray-based comparative genomic hybridization (CGH) panel on which BAC clones from chromosomes 13, 18, 21, X, and Y were spotted. Microarray-CGH analysis was performed for a total of 13 fetuses with congenital anomalies using cffDNA from their uncultured amniotic fluid. Microarray CGH with cffDNA led to successful molecular karyotyping for 12 of 13 fetuses within 5 days. Karyotypes of the 12 fetuses (one case of trisomy 13, two of trisomy 18, two of trisomy 21, one of monosomy X, and six of normal karyotype) were later confirmed by conventional chromosome analysis using cultured amniocytes. The one fetus whose molecular-karyotype was indicated as normal by microarray CGH actually had a balanced translocation, 45,XY,der(14;21)(q10;q10). The results indicated that microarray CGH with cffDNA is a useful rapid prenatal diagnostic method at late gestation for chromosome abnormalities with copy-number changes, especially when combined with conventional karyotyping of cultured amniocytes.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Amiel A, Bouaron N, Kidron D, Sharony R, Gaber E, Fejgin MD (2002) CGH in the detection of confined placental mosaicism (CPM) in placentas of abnormal pregnancies. Prenat Diagn 22:752–758
Barrett IJ, Lomax BL, Loukianova T, Tang SS, Lestou VS, Kalousek DK (2001) Comparative genomic hybridization: a new tool for reproductive pathology. Arch Pathol Lab Med 125:81–84
Bianchi DW (2000) Fetal cells in the mother: from genetic diagnosis to diseases associated with fetal cell microchimerism. Eur J Obstet Gynecol Reprod Biol 92:103–108
Bianchi DW, LeShane ES, Cowan JM (2001) Large amounts of cell-free fetal DNA are present in amniotic fluid. Clin Chem 47:1867–1869
Bianchi DW, Lo YM (2001) Fetomaternal cellular and plasma DNA trafficking: the Yin and the Yang. Ann NY Acad Sci 945:119–131
Drummond CL, Gomes DM, Senat MV, Audibert F, Dorion A, Ville Y (2003) Prenat. Diagn 23:1068–1072
Harada N, Hatchwell E, Okamoto N, Tsukahara M, Kurosawa K, Kawame H, Kondoh T, Ohashi H, Tsukino R, Kondoh Y, Shimokawa O, Ida T, Nagai T, Fukushima Y, Yoshiura K, Niikawa N, Matsumoto N (2004) Subtelomere specific microarray based comparative genomic hybridisation: a rapid detection system for cryptic rearrangements in idiopathic mental retardation. J Med Genet 41:130–136
Hahn S, Huppertz B, Holzgreve W (2005) Fetal cells and cell free fetal nucleic acids in maternal blood: new tools to study abnormal placentation? Placenta 26:515–526
Lapierre JM, Cacheux V, Luton D, Collot N, Oury JF, Aurias A, Tachdjian G (2000) Analysis of uncultured amniocytes by comparative genomic hybridization: a prospective prenatal study. Prenat Diagn 20:123–131
Larrabee PB, Johnson KL, Pestova E, Lucas M, Wilber K, LeShane ES, Tantravahi U, Cowan JM, Bianchi DW (2004) Microarray analysis of cell-free fetal DNA in amniotic fluid: a prenatal molecular karyotype. Am J Hum Genet 75:485–491
Lau TK, Leung TN (2005) Genetic screening and diagnosis. Curr Opin Obstet Gynecol 17:163–169
Lo YM (2005) Recent advances in fetal nucleic acids in maternal plasma. J Histochem Cytochem 53:293–296
Mantripragada KK, Buckley PG, Benetkiewicz M, de Bustos C, Hirvela C, Jarbo C, Bruder CEG, Wensman H, Mathiesen T, Nyberg G, Papi L, Collins VP, Ichimura K, Evans G, Dumanski JP (2003) High-resolution profiling of an 11 Mb segment of human chromosome 22 in sporadic schwannoma using array-CGH. Inter J Oncol 22:615–622
Masuzaki H, Miura K, Yoshimura S, Yoshiura K, Ishimaru T (2004) Detection of cell free placental DNA in maternal plasma: direct evidence from three cases of confined placental mosaicism. J Med Genet 41:289–292
Miyake N, Shimokawa O, Harada N, Sosonkina N, Okubo A, Kawara H, Okamoto N, Kurosawa K, Kawame H, Iwakoshi M, Kosho T, Fukushima Y, Makita Y, Yokoyama Y, Yamagata T, Kato M, Hiraki Y, Nomura M, Yoshiura K, Kishino T, Ohta T, Mizuguchi T, Niikawa N, Matsumoto N (2006) BAC array CGH reveals genomic aberrations in idiopathic mental retardation. Am J Med Genet 140:205–211
Nyberg DA, Souter VL (2000) Sonographic markers of fetal aneuploidy. Clin Perinatol 27:761–789
Oostlander AE, Meijer GA, Ylstra B (2004) Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet 66:488–495
Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO (1999) Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet 23:41–46
Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A (1992) Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics 13:718–725
Vermeesch JR, Melotte C, Froyen G, Van Vooren S, Dutta B, Maas N, Vermeulen S, Menten B, Speleman F, De Moor B, Van Hummelen P, Marynen P, Fryns JP, Devriendt K (2005) Molecular karyotyping: array CGH quality criteria for constitutional genetic diagnosis. J Histochem Cytochem 53:413–422
Wataganara T, Bianchi DW (2004) Fetal cell-free nucleic acids in the maternal circulation: new clinical applications. Ann NY Acad Sci 1022:90–99
Yang JH, Chung JH, Shin JS, Choi JS, Ryu HM, Kim MY (2005) Prenatal diagnosis of trisomy 18: report of 30 cases. Prenat Diagn 25:119–122
Acknowledgments
We thank Dr Joseph Wagstaff for his help and valuable advice. K.M., H.M., N.M., and N.N. were supported in part by Grants-in-Aid for Scientific Research (Nos. 16591670, 17591748, 16390101, and 13854024, respectively) from the Ministry of Education, Sports, Culture, Science and Technology of Japan, and N.N. was supported by SORST from Japan Science and Technology Agency (JST).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Miura, S., Miura, K., Masuzaki, H. et al. Microarray comparative genomic hybridization (CGH)-based prenatal diagnosis for chromosome abnormalities using cell-free fetal DNA in amniotic fluid. J Hum Genet 51, 412–417 (2006). https://doi.org/10.1007/s10038-006-0376-7
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1007/s10038-006-0376-7
Keywords
This article is cited by
-
Noninvasive fetal trisomy detection by multiplexed semiconductor sequencing: a barcoding analysis strategy
Journal of Human Genetics (2016)
-
Mosaic pregnancy after transfer of a “euploid” blastocyst screened by DNA microarray
Journal of Ovarian Research (2013)
-
Proteomic profile determination of autosomal aneuploidies by mass spectrometry on amniotic fluids
Proteome Science (2008)
-
Guidelines for molecular karyotyping in constitutional genetic diagnosis
European Journal of Human Genetics (2007)
-
Identification of origin of unknown derivative chromosomes by array-based comparative genomic hybridization using pre- and postnatal clinical samples
Journal of Human Genetics (2007)