Abstract
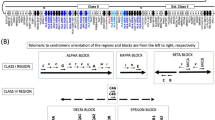
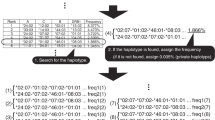
The human leukocyte antigen (HLA) complex locus has shaped a framework for evolutionary processes because of the dense clustering and strong linkage disequilibrium (LD) of polymorphic genes. Although the landscape of LD among conventional single-nucleotide polymorphisms (SNPs) has been described, the data on the lineage of major histocompatibility complex (MHC) haplotype are limited to pairwise comparisons of several haplotypes in Caucasoid populations. Multi-allelic markers, including microsatellite markers, may provide us with a larger power to analyze the MHC haplotype lineage because the mutation rate of microsatellite exceeds that of SNPs by several orders of magnitude. In this study, we investigated the complex structure of repeat motifs in a microsatellite to figure out the structural lineage of HLA-Cw/-B segments in Japanese. It was found that the genetic differences of HLA-Cw/-B haplotype lineage were reflected by repeat motif patterns at C1_2_5 locus, suggesting that unique mutational dynamics of microsatellites may be a useful marker to chase the haplotype lineage.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Horton, R., Wilming, L., Rand, V., Lovering, R. C., Bruford, E. A., Khodiyar, V. K. et al. Gene map of the extended human MHC. Nat. Rev. Genet. 5, 889–899 (2004).
Cooke, G. S. & Hill, A. V. Genetics of susceptibility to human infectious disease. Nat. Rev. Genet. 2, 967–977 (2001).
Rudolph, M. G., Stanfield, R. L. & Wilson, I. A. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 (2006).
Little, A. M. & Parham, P. Polymorphism and evolution of HLA class I and II genes and molecules. Rev. Immunogenet. 1, 105–123 (1999).
Meyer, D., Single, R. M., Mack, S. J., Erlich, H. A. & Thomson, G. Signatures of demographic history and natural selection in the human major histocompatibility complex loci. Genetics. 173, 2121–2142 (2006).
Miretti, M. M., Walsh, E. C., Ke, X., Delgado, M., Griffiths, M., Hunt, S. et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 76, 634–646 (2005).
Shichi, D., Kikkawa, E. F., Ota, M., Katsuyama, Y., Kimura, A., Matsumori, A. et al. The haplotype block, NFKBIL1-ATP6V1G2-BAT1-MICB-MICA, within the class III-class I boundary region of the human major histocompatibility complex may control susceptibility to hepatitis C virus-associated dilated cardiomyopathy. Tissue Antigens 66, 200–208 (2005).
Shiina, T., Ota, M., Shimizu, S., Katsuyama, Y., Hashimoto, N., Takasu, M. et al. Rapid evolution of major histocompatibility complex class I genes in primates generates new disease alleles in humans via hitchhiking diversity. Genetics 173, 1555–1570 (2006).
Ahmad, T., Neville, M., Marshall, S. E., Armuzzi, A., Mulcahy-Hawes, K., Crawshaw, J. et al. Haplotype-specific linkage disequilibrium patterns define the genetic topography of the human MHC. Hum. Mol. Genet. 12, 647–656 (2003).
Horton, R., Gibson, R., Coggill, P., Miretti, M., Allcock, R. J., Almeida, J. et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 60, 1–18 (2008).
Nagy, M., Entz, P., Otremba, P., Schoenemann, C., Murphy, N. & Dapprich, J. Haplotype-specific extraction: a universal method to resolve ambiguous genotypes and detect new alleles—demonstrated on HLA-B. Tissue Antigens 69, 176–180 (2007).
Subirana, J. A. & Messeguer, X. Structural families of genomic microsatellites. Gene 408, 124–132 (2008).
Matsuzaka, Y., Makino, S., Nakajima, K., Tomizawa, M., Oka, A., Bahram, S. et al. New polymorphic microsatellite markers in the human MHC class III region. Tissue Antigens 57, 397–404 (2001).
Matsuzaka, Y., Makino, S., Nakajima, K., Tomizawa, M., Oka, A., Kimura, M. et al. New polymorphic microsatellite markers in the human MHC class II region. Tissue Antigens 56, 492–500 (2000).
Foissac, A., Salhi, M. & Cambon-Thomsen, A. Microsatellites in the HLA region: 1999 update. Tissue Antigens 55, 477–509 (2000).
Cullen, M., Malasky, M., Harding, A. & Carrington, M. High-density map of short tandem repeats across the human major histocompatibility complex. Immunogenetics 54, 900–910 (2003).
Malkki, M., Single, R., Carrington, M., Thomson, G. & Petersdorf, E. MHC microsatellite diversity and linkage disequilibrium among common HLA-A, HLA-B, DRB1 haplotypes: implications for unrelated donor hematopoietic transplantation and disease association studies. Tissue Antigens 66, 114–124 (2005).
Mungall, A. J., Palmer, S. A., Sims, S. K., Edwards, C. A., Ashurst, J. L., Wilming, L. et al. The DNA sequence and analysis of human chromosome 6. Nature 425, 805–811 (2003).
Saito, S., Ota, S., Yamada, E., Inoko, H. & Ota, M. Allele frequencies and haplotypic associations defined by allelic DNA typing at HLA class I and class II loci in the Japanese population. Tissue Antigens 56, 522–529 (2000).
Stewart, C. A., Horton, R., Allcock, R. J., Ashurst, J. L., Atrazhev, A. M., Coggill, P. et al. Complete MHC haplotype sequencing for common disease gene mapping. Genome Res. 14, 1176–1187 (2004).
Lancaster, A. K., Single, R. M., Solberg, O. D., Nelson, M. P. & Thomson, G. PyPop update-software pipeline for large-scale multilocus population genomics. Tissue Antigens 69 (Suppl 1), 192–197 (2007).
Warnes, G. R. The genetics package. R News 3, 9–13 (2003).
Schaid, D. J., Rowland, C. M., Tines, D. E., Jacobson, R. M. & Poland, G. A. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am. J. Hum. Genet. 70, 425–434 (2002).
Sabeti, P. C., Reich, D. E., Higgins, J. M., Levine, H. Z., Richter, D. J., Schaffner, S. F. et al. Detecting recent positive selection in the human genome from haplotype structure. Nature 419, 832–837 (2002).
Hedrick, P. W. Gametic disequilibrium measures: proceed with caution. Genetics 117, 331–341 (1987).
Cramer, H. Mathematical Methods of Statistics (Princeton University Press, Princeton, New Jersey, 1946).
Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Tokunaga, K., Ishikawa, Y., Ogawa, A., Wang, H., Mitsunaga, S., Moriyama, S. et al. Sequence-based association analysis of HLA class I and II alleles in Japanese supports conservation of common haplotypes. Immunogenetics 46, 199–205 (1997).
Ellegren, H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 5, 435–445 (2004).
Lai, Y. & Sun, F. The relationship between microsatellite slippage mutation rate and the number of repeat units. Mol. Biol. Evol. 20, 2123–2131 (2003).
Boyer, J. C., Hawk, J. D., Stefanovic, L. & Farber, R. A. Sequence-dependent effect of interruptions on microsatellite mutation rate in mismatch repair-deficient human cells. Mutat. Res. 640, 89–96 (2008).
Macaubas, C., Jin, L., Hallmayer, J., Kimura, A. & Mignot, E. The complex mutation pattern of a microsatellite. Genome Res. 7, 635–641 (1997).
Leslie, S., Donnelly, P. & McVean, G. A Statistical method for predicting classical HLA alleles from SNP data. Am. J. Hum. Genet. 82, 48–56 (2008).
de Bakker, P. I., McVean, G., Sabeti, P. C., Miretti, M. M., Green, T., Marchini, J. et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 38, 1166–1172 (2006).
Acknowledgements
We are grateful to Atsuko Shigenari, Eri F Kikkawa, Hisako Kawata, Norihisa Sugita, Daisuke Sumiyama, Ryoko Ohkubo, Rui-Ping Dong, Shuji Hoshino, Masayuki Yoshida, Shin-ichiro Yasunaga and Yukiji Date for their contributions in genotyping. This work was supported in part by Grant-in-Aids from the Ministry of Education, Science, Sports, Culture and Technology (MEXT) of Japan, the program of Founding Research Centers for Emerging and Reemerging Infection Disease supported by MEXT, Japan, and research grants from the Ministry of Health, Labor and Welfare, Japan, and from the Japan Health Sciences Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Shichi, D., Ota, M., Katsuyama, Y. et al. Complex divergence at a microsatellite marker C1_2_5 in the lineage of HLA-Cw/-B haplotype. J Hum Genet 54, 224–229 (2009). https://doi.org/10.1038/jhg.2009.15
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2009.15