Abstract
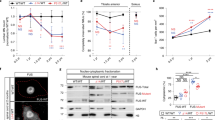
Amyotrophic lateral sclerosis (ALS) is a severe neurodegenerative disease characterized by the degeneration of upper and lower motor neurons. Genetic studies have led, thus far, to the identification of 12 loci and 9 genes for familial ALS (FALS). Although the distribution and impact of superoxide dismutase 1 mutations has been extensively examined for over a decade, the recently identified FALS-associated FUS gene has been less studied. Therefore, we set out to screen our collection of FALS cases for FUS mutations. All 15 exons of FUS were amplified and sequenced in 154 unrelated FALS cases and 475 ethnically matched healthy individuals. One substitution located in the acceptor splice site of intron 14 was identified in all affected members of a large family, causing the skipping of the last 13 amino acids of the protein and the translation of 7 novel amino acids, resulting from the new translation of a part of the 3′ untranslated region. Our study identified a new splicing mutation in the highly conserved C-terminal of the FUS protein. Thus far most FUS mutations are missenses, and our findings, combined with those of others, confirm the importance of the C-terminal portion of the protein, adding additional support for FUS mutations having a critical role in ALS.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Shaw, C. E., Enayat, Z. E., Powell, J. F., Anderson, V. E., Radunovic, A., Al-Sarraj, S. et al. Familial amyotrophic lateral sclerosis. Molecular pathology of a patient with a SOD1 mutation. Neurology 49, 1612–1616 (1997).
Camu, W., Khoris, J., Moulard, B., Salachas, F., Briolotti, V., Rouleau, G. A. et al. Genetics of familial ALS and consequences for diagnosis. French ALS Research Group. J. Neurol. Sci. 165 (Suppl 1), S21–S26 (1999).
Kurtzke, J. F. Epidemiology of amyotrophic lateral sclerosis. Adv. Neurol. 36, 281–302 (1982).
Rosen, D. R., Siddique, T., Patterson, D., Figlewicz, D. A., Sapp, P., Hentati, A. et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 (1993).
Sreedharan, J., Blair, I. P., Tripathi, V. B., Hu, X., Vance, C., Rogelj, B. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008).
Kwiatkowski, T. J. Jr, Bosco, D. A., Leclerc, A. L., Tamrazian, E., Vanderburg, C. R., Russ, C. et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323, 1205–1208 (2009).
Vance, C., Rogelj, B., Hortobagyi, T., De Vos, K. J., Nishimura, A. L., Sreedharan, J. et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323, 1208–1211 (2009).
Lagier-Tourenne, C. & Cleveland, D. W. Rethinking ALS: the FUS about TDP-43. Cell 136, 1001–1004 (2009).
Daoud, H., Valdmanis, P. N., Kabashi, E., Dion, P., Dupre, N., Camu, W. et al. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J. Med. Genet. 46, 112–114 (2009).
Valdmanis, P. N., Belzil, V. V., Lee, J., Dion, P. A., St-Onge, J., Hince, P. et al. A mutation that creates a pseudoexon in SOD1 causes familial ALS. Ann. Hum. Genet. 73, 652–657 (2009).
Zu, J. S., Deng, H. X., Lo, T. P., Mitsumoto, H., Ahmed, M. S., Hung, W. Y. et al. Exon 5 encoded domain is not required for the toxic function of mutant SOD1 but essential for the dismutase activity: identification and characterization of two new SOD1 mutations associated with familial amyotrophic lateral sclerosis. Neurogenetics 1, 65–71 (1997).
Dejesus-Hernandez, M., Kocerha, J., Finch, N., Crook, R., Baker, M., Desaro, P. et al. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum. Mutat. 31, 1377–1389 (2010).
Nlend, R., Meyer, K. & Schumperli, D. Repair of pre-mRNA splicing: Prospects for a therapy for Spinal Muscular Atrophy. RNA Biol. 7, 430–440 (2010).
Pros, E., Fernandez-Rodriguez, J., Benito, L., Ravella, A., Capella, G., Blanco, I. et al. Modulation of aberrant NF1 pre-mRNA splicing by kinetin treatment. Eur. J. Hum. Genet. 18, 614–617 (2009).
O'Leary, D. A., Vargas, L., Sharif, O., Garcia, M. E., Sigal, Y. J., Chow, S. K. et al. HTS-compatible patient-derived cell-based assay to identify small molecule modulators of aberrant splicing in myotonic dystrophy type 1. Curr. Chem. Genomics. 4, 9–18 (2010).
Acknowledgements
VVB, HD and GAR are supported by the Canadian Institutes of Health Research. We would like to thank the patients involved in this study; Mélanie Benard, Isabelle Thibault and Pierre Provencher for sample collection and organization; Anne Noreau, Cynthia Bourassa, Sophie Massart and Bertrand Boutié for technical support; and to acknowledge support from the Association pour la Recherche sur la Sclérose Latérale Amyotrophique (ARS), the Association Française contre les Myopathies (AFM) and the French Group on MND.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Belzil, V., St-Onge, J., Daoud, H. et al. Identification of a FUS splicing mutation in a large family with amyotrophic lateral sclerosis. J Hum Genet 56, 247–249 (2011). https://doi.org/10.1038/jhg.2010.162
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2010.162
Keywords
This article is cited by
-
Pur-alpha regulates cytoplasmic stress granule dynamics and ameliorates FUS toxicity
Acta Neuropathologica (2016)