Abstract
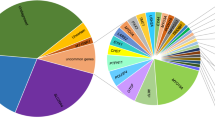
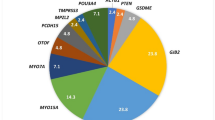
Hearing loss (HL) is the most prevalent sensory defect affecting 1 in 500 neonates. Genetic factors are involved in half of the cases. The extreme heterogeneity of HL makes it difficult to analyze and determine the accurate genetic causes of the impairment. Up to now, 10 genes, namely, GJB2, GJB6, SLC26A4, TECTA, PJVK, Col11A2, Myo15A, TMC1, RDX and microRNA (miR-183), have been studied in an Iranian population. The prevalence of HL in Iran was estimated to be 2–3 times higher than that in other parts of the world. Here, the most common bases of congenital nonsyndromic hearing loss (NSHL) are discussed. We reviewed GJB2, GJB6 (large deletion), TECTA, SLC26A4 and PEJVK mutations, and studied their frequencies and distributions in different ethnic groups in 1934, 500, 121, 80 and 34 unrelated families throughout Iran, respectively. GJB2 mutation was the most common factor causing NSHL, with a mean frequency of 18.17% in the Iranian population. The importance of Iran's geographical location in the migration pathway from west to east through the silk route was also highlighted. SLC26A4 and TECTA mutations were the second and third main reasons of HL and accounted for up to 10 and 4% of prelingual HL in Iran, respectively. Mutations in GJB2, SLC26, TECTA and PJVK genes have an important role in HL in Iran and a screening test should be generated for better intervention and diagnosis programs.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Morton, C. C. & Nance, W. E. Newborn hearing screening--a silent revolution. N. Engl. J. Med. 354, 2151–2164 (2006).
Hilgert, N., Alasti, F., Dieltjens, N., Pawlik, B., Wollnik, B. & Uyguner, O. Mutation analysis of TMC1 identifies four new mutations and suggests an additional deafness gene at loci DFNA36 and DFNB7/11. Clin. Genet. 74, 223–232 (2008).
Dror, A. A. & Avraham, K. B. Hearing loss: mechanisms revealed by genetics and cell biology. Annu. Rev. Genet. 43, 411–437 (2009).
Mencía, A., Modamio-Høybjør, S., Redshaw, N., Morín, M., Mayo-Merino, F., Olavarrieta, L. et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 41, 609–613 (2009).
Morton, N. E. Genetic epidemiology of hearing impairment. Ann. NY. Acad. Sci. 630, 16–31 (1991).
Cryns, K. & van Camp, G. Deafness genes and their diagnostic applications. Audiol. Neurootol. 9, 2–22 (2004).
Finsterer, J. & Fellinger, J. Nuclear and mitochondrial genes mutated in nonsyndromic impaired hearing. Int. J. Pediatr. Otorhinolaryngol. 69, 621–647 (2005).
Quds Newspaper. http://www.qudsdaily.com/archive/1388/html/7/1388-07-07/page2.html (2009).
Cotton, R. G., Al Aqeel, A. I., Al-Mulla, F., Carrera, P., Claustres, M., Ekong, R. et al. Capturing all disease-causing mutations for clinical and research use: toward an effortless system for the Human Variome Project. Genet. Med. 11, 843–849 (2009).
Ben-Yosef, T. & Friedman, T. B. The genetic bases for syndromic and nonsyndromic deafness among Jews. Trends Mol. Med. 9, 496–502 (2003).
Usami, S., Wagatsuma, M., Fukuoka, H., Suzuki, H., Tsukada, K., Nishio, S. et al. The responsible genes in Japanese deafness patients and clinical application using Invader assay. Acta. Otolaryngol. 128, 446–454 (2008).
Ouyang, X. M., Yan, D., Yuan, H. J., Pu, D., Du, L. L., Han, D. Y. et al. The genetic bases for non-syndromic hearing loss among Chinese. J. Hum. Genet. 54, 131–140 (2009).
Guilford, P., Ben Arab, S., Blanchard, S., LeVilliers, J., Weissenbach, J., Belkahia, A. et al. A non-syndrome form of neurosensory, recessive deafness maps to the pericentromeric region of chromosome 13q. Nat. Genet. 6, 24–28 (1994).
Goodenough, D. A., Goliger, J. A. & Paul, D. L. Connexins, connexons, and intercellular communication. Annu. Rev. Biochem. 65, 475–502 (1996).
Tekin, M., Arnos, K. S. & Pandya, A. Advances in hereditary deafness. Lancet 358, 1082–1090 (2001).
Mahdieh, N. & Rabbani, B. Statistical study of 35delG mutation of GJB2 gene: a meta-analysis of carrier frequency. Int. J. Audiol. 48, 363–370 (2009).
Morell, R. J., Kim, H. J., Hood, L. J., Goforth, L., Friderici, K., Fisher, R. et al. Mutations in the connexin 26 gene (GJB2) among Ashkenazi Jews with nonsyndromic recessive deafness. N. Engl. J. Med. 339, 1500–1505 (1998).
Kudo, T., Ikeda, K., Kure, S., Matsubara, Y., Oshima, T., Watanabe, K. et al. Novel mutations in the connexin 26 gene (GJB2) responsible for childhood deafness in the Japanese population. Am. J. Med. Genet. 90, 141–145 (2000).
Brobby, G. W., Muller-Myhsok, B. & Horstmann, R. D. Connexin 26 R143W mutation associated with recessive nonsyndromic sensorineural deafness in Africa. N. Engl. J. Med. 338, 548–549 (1998).
Hamelmann, C., Amedofu, G. K., Albrecht, K., Muntau, B., Gelhaus, A., Brobby, G. W. et al. Pattern of connexin 26 (GJB2) mutations causing sensorineural hearing impairment in Ghana. Hum. Mutat. 18, 84–85 (2001).
Maheshwari, M., Vijaya, R., Ghosh, M., Shastri, S., Kabra, M. & Menon, P. S. Screening of families with autosomal recessive non-syndromic hearing impairment (ARNSHI) for mutations in GJB2 gene: Indian scenario. Am. J. Med. Genet. A. 120, 180–184 (2003).
Bouwer, S., Angelicheva, D., Chandler, D., Seeman, P., Tournev, I. & Kalaydjieva, L. Carrier rates of the ancestral Indian W24X mutation in GJB2 in the general Gypsy population and individual subisolates. Genet. Test. 11, 455–458 (2007).
Mahdieh, N., Bagherian, H., Shirkavand, A., Sharafi, M. & Zeinali, S. High level of intrafamilial phenotypic variability of non-syndromic hearing loss in a Lur family due to delE120 mutation in GJB2 gene. Int. J. Pediatr. Otorhinolaryngol. 74, 1089–1091 (2010).
Najmabadi, H., Nishimura, C., Kahrizi, K., Riazalhosseini, Y., Malekpour, M., Daneshi, A. et al. GJB2 mutations: passage through Iran. Am. J. Med. Genet. A 133A, 132–137 (2005).
Hashemzadeh Chaleshtori, M., Farhud, D. D. & Patton, M. A. Familial and sporadic GJB2-related deafness in Iran: review of gene mutations. Iran. J. Public Health 36, 1–14 (2007).
Bonyadi, M., Esmaeili, M., Abhari, M. & Lotfi, A. Mutation analysis of familial GJB2-related deafness in Iranian Azeri, Turkish patients. Genet. Test. Mol. Biomarkers 13, 689–692 (2009).
Mahdieh, N., Nishimura, C., Ali-Madadi, K., Riazalhosseini, Y., Yazdan, H., Arzhangi, S. et al. The frequency of GJB2 mutations and the (GJB6-D13S1830) deletion as a cause of autosomal recessive non-syndromic deafness in the Kurdish population. Clin. Genet. 65, 506–508 (2004).
Kelley, P. M., Harris, D. J., Comer, B. C., Askew, J. W., Fowler, T., Smith, S. D. et al. Novel mutations in the connexin26 gene (GJB2) that cause autosomal recessive (DFNB1) hearing loss. Am. J. Hum. Genet. 62, 792–799 (1998).
Abe, S., Usami, S., Shinkawa, H., Kelley, P. M. & Kimberling, W. J. Prevalent connexin 26 gene (GJB2) mutations in Japanese. J. Med. Genet. 37, 41–43 (2000).
Lee, K. Y., Choi, S. Y., Bae, J. W., Kim, S., Chung, K. W., Drayna, D. et al. Molecular analysis of the GJB2, GJB6 and SLC26A4 genes in Korean deafness patients. Int. J. Pediatr. Otorhinolaryngol. 72, 1301–1309 (2008).
Dai, P., Yu, F., Han, B., Yuan, Y., Li, Q., Wang, G. et al. The prevalence of the 235delC GJB2 mutation in a Chinese deaf population. Genet. Med. 9, 283–289 (2007).
Dai, P., Yu, F., Han, B., Liu, X., Wang, G., Li, Q. et al. GJB2 mutation spectrum in 2,063 Chinese patients with nonsyndromic hearing impairment. J. Transl. Med. 7, 26 (2009).
Roux, A. F., Pallares-Ruiz, N., Vielle, A., Faugère, V., Templin, C., Leprevost, D. et al. Molecular epidemiology of DFNB1 deafness in France. BMC Med. Genet. 5, 5 (2004).
Liu, X. Z., Yuan, Y., Yan, D., Ding, E. H., Ouyang, X. M., Fei, Y. et al. Digenic inheritance of non-syndromic deafness caused by mutations at the gap junction proteins Cx26 and Cx31. Hum. Genet. 125, 53–62 (2009).
Lamartine, J., Munhoz Essenfelder, G., Kibar, Z., Lanneluc, I., Callouet, E., Laoudj, D. et al. Mutations in GJB6 cause hidrotic ectodermal dysplasia. Nat. Genet. 26, 142–144 (2000).
Lerer, I., Sagi, M., Ben-Neriah, Z., Wang, T., Levi, H. & Abeliovich, D. A deletion mutation in GJB6 cooperating with a GJB2 mutation in trans in non-syndromic deafness: A novel founder mutation in Ashkenazi Jews. Hum. Mutat. 18, 460 (2001).
Pallares-Ruiz, N., Blanchet, P., Mondain, M., Claustres, M. & Roux, A. F. A large deletion including most of GJB6 in recessive non syndromic deafness: a digenic effect? Eur. J. Hum. Genet. 10, 72–76 (2002).
del Castillo, I., Villamar, M., Moreno-Pelayo, M A., del Castillo, F. J., Alvarez, A., Tellería, D. et al. A deletion involving the connexin 30 gene in nonsyndromic hearing impairment. N. Engl. J. Med. 346, 243–249 (2002).
del Castillo, F. J., Rodríguez-Ballesteros, M., Alvarez, A., Hutchin, T., Leonardi, E., de Oliveira, C. A. et al. A novel deletion involving the connexin-30 gene, del(GJB6-d13s1854), found in trans with mutations in the GJB2 gene (connexin-26) in subjects with DFNB1 non-syndromic hearing impairment. J. Med. Genet. 42, 588–594 (2005).
Feldmann, D., Le Maréchal, C., Jonard, L., Thierry, P., Czajka, C., Couderc, R. et al. A new large deletion in the DFNB1 locus causes nonsyndromic hearing loss. Eur. J. Med. Genet. 52, 195–200 (2009).
Wilch, E., Azaiez, H., Fisher, R. A., Elfenbein, J., Murgia, A., Birkenhäger, R. et al. A novel DFNB1 deletion allele supports the existence of a distant cis-regulatory region that controls GJB2 and GJB6 expression. Clin. Genet. 78, 267–274 (2010).
del Castillo, I., Moreno-Pelayo, M. A., del Castillo, F. J., Brownstein, Z., Marlin, S., Adina, Q. et al. Prevalence and evolutionary origins of the del(GJB6-D13S1830) mutation in the DFNB1 locus in hearing impaired subjects: a multicenter study. Am. J. Hum. Genet. 73, 1452–1458 (2003).
Marlin, S., Feldmann, D., Blons, H., Loundon, N., Rouillon, I., Albert, S. et al. GJB2 and GJB6 mutations: genotypic and phenotypic correlations in a large cohort of hearing-impaired patients. Arch. Otolaryngol. Head Neck Surg. 131, 481–487 (2005).
Gravina, L. P., Foncuberta, M. E., Prieto, M. E., Garrido, J., Barreiro, C. & Chertkoff, L. Prevalence of DFNB1 mutations in Argentinean children with non-syndromic deafness. Report of a novel mutation in GJB2. Int. J. Pediatr. Otorhinolaryngol. 74, 250–254 (2010).
Batissoco, A. C., Abreu-Silva, R. S., Braga, M. C., Lezirovitz, K., Della-Rosa, V., Alfredo, T. Jr. et al. Prevalence of GJB2 (connexin-26) and GJB6 (connexin-30) mutations in a cohort of 300 Brazilian hearing-impaired individuals: implications for diagnosis and genetic counseling. Ear Hear. 30, 1–7 (2009).
Riazalhosseini, Y., Nishimura, C., Kahrizi, K., Shafeghati, Y., Danseshi, A., Jogataie, M. T. et al. Delta (GJB6-D13S1830) is not a common cause of nonsyndromic hearing loss in in the Iranian patients. Arch. Iran. Med. 8, 104–108 (2005).
Esmaeili, M., Bonyadi, M. & Nejadkazem, M. Common mutation analysis of GJB2 and GJB6 genes in affected families with autosomal recessive non-syndromic hearing loss from Iran: simultaneous detection of two common mutations (35delG=del(GJB6-D13S1830)) in the DFNB1-related deafness. Int. J. Pediatr. Otorhinolaryngol. 71, 869–873 (2007).
Evirgen, N., Solak, M., Dereköy, S., Erdo∂an, M., Yildiz, H., Eser, B. et al. Genotyping for Cx26 and Cx30 mutations in cases with congenital hearing loss. Genet. Test. 12, 253–256 (2008).
Padma, G., Ramchander, P. V., Nandur, U. V. & Padma, T. GJB2 and GJB6 gene mutations found in Indian probands with congenital hearing impairment. J. Genet. 88, 267–272 (2009).
Sansoviæ, I., Knezeviæ, J., Musani, V., Seeman, P., Barisiæ, I. & Paveliæ, J. GJB2 mutations in patients with nonsyndromic hearing loss from Croatia. Genet. Test. Mol. Biomarkers 13, 693–699 (2009).
Sheffield, V. C., Kraiem, Z., Beck, J. C., Nishimura, D., Stone, E. M., Salameh, M. et al. Pendred syndrome maps to chromosome 7q21-34 and is caused by an intrinsic defect in thyroid iodine organification. Nat. Genet. 12, 424–426 (1996).
Borck, G., Roth, C., Martiné, U., Wildhardt, G. & Pohlenz, J. Mutations in the PDS gene in German families with Pendred′s syndrome: V138F is a founder mutation. J. Clin. Endocrinol. Metab. 88, 2916–2921 (2003).
Gillam, M. P., Sidhaye, A. R., Lee, E. J., Rutishauser, J., Stephan, C. W. & Kopp, P. Functional characterization of pendrin in a polarized cell system. Evidence for pendrin-mediated apical iodide efflux. J. Biol. Chem. 279, 13004–13010 (2004).
Dossena, S., Rodighiero, S., Vezzoli, V., Nofziger, C., Salvioni, E., Boccazzi, M. et al. Functional characterization of wild-type and mutated pendrin (SLC26A4), the anion transporter involved in Pendred syndrome. J. Mol. Endocrinol. 43, 93–103 (2009).
Mount, D. B. & Romero, M. F. The SLC26 gene family of multifunctional anion exchangers. Pflugers Arch. 447, 710–721 (2004).
Everett, L. A., Glaser, B., Beck, J. C., Idol, J. R., Buchs, A., Heyman, M. et al. Pendred syndrome is caused by mutations in a putative sulphate transporter gene (PDS). Nat. Genet. 17, 411–422 (1997).
Li, X. C., Everett, L. A., Lalwani, A. K., Desmukh, D., Friedman, T. B., Green, E. D. et al. A mutation in PDS causes non-syndromic recessive deafness. Nat. Genet. 18, 215–217 (1998).
Campbell, C., Cucci, R. A., Prasad, S., Green, G. E., Edeal, J. B., Galer, C. E. et al. Pendred syndrome, DFNB4, and PDS/SLC26A4 identification of eight novel mutations and possible genotype-phenotype correlations. Hum. Mutat. 17, 403–411 (2001).
Smith, R. I. H. & Hone, S. Genetic screening for deafness. Pediatr. Clin. North Am. 50, 315–329 (2003).
Kahrizi, K., Mohseni, M., Nishimura, C., Bazazzadegan, N., Fischer, S. M., Dehghani, A. et al. Identification of SLC26A4 gene mutations in Iranian families with hereditary hearing impairment. Eur. J. Pediatr. 168, 651–653 (2009).
Schrijver, I. Hereditary non-syndromic sensorineural hearing loss: transforming silence to sound. J. Mol. Diagn. 6, 275–284 (2004).
Harada, D., Namba, A., Abe, S. & Usami, S Distribution and frequencies of PDS (SLC26A4) mutations in Pendred syndrome and nonsyndromic hearing loss associated with enlarged vestibular aqueduct: a unique spectrum of mutations in Japanese. Eur. J. Hum. Genet. 11, 916–922 (2003).
Usami, S., Abe, S., Weston, M. D., Shinkawa, H., Van Camp, G. & Kimberling, W. J. Non-syndromic hearing loss associated with enlarged vestibular aqueduct is caused by PDS mutations. Hum. Genet. 104, 188–192 (1999).
Wang, Q. J., Zhao, Y. L., Rao, S. Q., Guo, Y. F., Yuan, H., Zong, L. et al. A distinct spectrum of SLC26A4 mutations in patients with enlarged vestibular aqueduct in China. Clin. Genet. 72, 245–254 (2007).
Hu, H., Wu, L., Feng, Y., Pan, Q., Long, Z., Li, J. et al. Molecular analysis of hearing loss associated with enlarged vestibular aqueduct in the Mainland Chinese: a unique SLC26A4 mutation spectrum. J. Hum. Genet. 52, 492–497 (2007).
Delmaghani, S., del Castillo, F. J., Michel, V., Leibovici, M., Aghaie, A., Ron, U. et al. Mutations in the gene encoding pejvakin, a newly identified protein of the afferent auditory pathway, cause DFNB59 auditory neuropathy. Nat. Genet. 38, 770–778 (2006).
Schwander, M., Sczaniecka, A., Grillet, N., Bailey, J. S., Avenarius, M., Najmabadi, H. et al. A forward genetics screen in mice identifies recessive deafness traits and reveals that pejvakin is essential for outer hair cell function. J. Neurosci. 27, 2163–2175 (2007).
Ebermann, I., Walger, M., Scholl, H. P., Charbel Issa, P., Lüke, C., Nürnberg, G. et al. Truncating mutation of the DFNB59 gene causes cochlear hearing impairment and central vestibular dysfunction. Hum. Mutat. 28, 571–577 (2007).
Hashemzadeh Chaleshtori, M., Simpson, M. A., Farrokhi, E., Dolati, M., Hoghooghi Rad, L., Amani Geshnigani, S. & Crosby, A. H. Novel mutations in the pejvakin gene are associated with autosomal recessive non-syndromic hearing loss in Iranian families. Clin. Genet. 72, 261–263 (2007).
Collin, R. W., Kalay, E., Oostrik, J., Caylan, R., Wollnik, B., Arslan, S. et al. Involvement of DFNB59 mutations in autosomal recessive nonsyndromic hearing impairment. Hum. Mutat. 28, 718–723 (2007).
Verhoeven, K., Van Laer, L., Kirschhofer, K., Legan, P. K., Hughes, D. C., Schatteman, I. et al. Mutations in the human alpha-tectorin gene cause autosomal dominant nonsyndromic hearing impairment. Nat. Genet. 19, 60–62 (1998).
Alloisio, N., Morlé, L., Bozon, M., Godet, J., Verhoeven, K., Van Camp, G. et al. Mutation in the zonadhesin-like domain of alpha-tectorin associated with autosomal dominant non-syndromic hearing loss. Eur. J. Hum. Genet. 7, 255–258 (1999).
Balciuniene, J., Dahl, N., Jalonen, P., Verhoeven, K., Van Camp, G., Borg, E. et al. Alpha-tectorin involvement in hearing disabilities: one gene--two phenotypes. Hum. Genet. 105, 211–216 (1999).
Mustapha, M., Weil, D., Chardenoux, S., Elias, S., El-Zir, E., Beckmann, J. S. et al. An alpha-tectorin gene defect causes a newly identified autosomal recessive form of sensorineural pre-lingual non-syndromic deafness, DFNB21. Hum. Mol. Genet. 8, 409–412 (1999).
Moreno-Pelayo, M. A., Goodyear, R. J., Mencía, A., Modamio-Høybjør, S., Legan, P. K., Olavarrieta, L. et al. Characterization of a spontaneous, recessive, missense mutation arising in the Tecta gene. J. Assoc. Res. Otolaryngol. 9, 202–214 (2008).
Naz, S., Alasti, F., Mowjoodi, A., Riazuddin, S., Sanati, M. H., Friedman, T. B. et al. Distinctive audiometric profile associated with DFNB21 alleles of TECTA. J. Med. Genet. 40, 360–363 (2003).
Meyer, N. C., Alasti, F., Nishimura, C. J., Imanirad, P., Kahrizi, K., Riazalhosseini, Y. et al. Identification of three novel TECTA mutations in Iranian families with autosomal recessive nonsyndromic hearing impairment at the DFNB21 locus. Am. J. Med. Genet. A 143A, 1623–1629 (2007).
Alasti, F., Sanati, M. H., Behrouzifard, A. H., Sadeghi, A., de Brouwer, A. P., Kremer, H. et al. A novel TECTA mutation confirms the recognizable phenotype among autosomal recessive hearing impairment families. Int. J. Pediatr. Otorhinolaryngol. 72, 249–255 (2008).
Legan, P. K., Rau, A., Keen, J. N. & Richardson, G. P. The mouse tectorins. Modular matrix proteins of the inner ear homologous to components of the sperm-egg adhesion system. J. Biol. Chem. 272, 8791–8801 (1997).
Pfister, M., Thiele, H., Van Camp, G., Fransen, E., Apaydin, F., Aydin, O. et al. A genotype-phenotype correlation with gender-effect for hearing impairment caused by tecta mutations. Cell. Physiol. Biochem. 14, 369–376 (2004).
Plantinga, R. F., de Brouwer, A. P., Huygen, P. L., Kunst, H. P., Kremer, H. & Cremers, C. W. A novel TECTA mutation in a Dutch DFNA8/12 family confirms genotype-phenotype correlation. J. Assoc. Res. Otolaryngol. 7, 173–181 (2006).
Collin, R. W., de Heer, A. M., Oostrik, J., Pauw, R. J., Plantinga, R. F., Huygen, P. L. et al. Mid-frequency DFNA8/12 hearing loss caused by a synonymous TECTA mutation that affects an exonic splice enhancer. Eur. J. Hum. Genet. 16, 1430–1436 (2008).
Greinwald, J. H. Jr., Scott, D. A., Marietta, J. R., Carmi, R., Manaligod, J., Ramesh, A. et al. Construction of P1-derived artificial chromosome and yeast artificial chromosome contigs encompassing the DFNB7 and DFNB11 region of chromosome 9q13-21. Genome Res. 7, 879–886 (1997).
Kurima, K., Peters, L. M., Yang, Y., Riazuddin, S., Ahmed, Z. M., Naz, S. et al. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet. 30, 277–284 (2002).
Kurima, K., Yang, Y., Sorber, K. & Griffith, A. J. Characterization of the transmembrane channellike (TMC) gene family: functional clues from hearing loss and epidermodysplasia verruciformis. Genomics 81, 300–308 (2003).
Kitajiri, S. I., McNamara, R., Makishima, T., Husnain, T., Zafar, A. U., Kittles, R. A. et al. Identities, frequencies and origins of TMC1 mutations causing DFNB7/B11 deafness in Pakistan. Clin. Genet. 72, 546–550 (2007).
Kalay, E., Karaguzel, A., Caylan, R., Heister, A., Cremers, F. P., Cremers, C. W. et al. Four novel TMC1 (DFNB7/DFNB11) mutations in Turkish patients with congenital autosomal recessive nonsyndromic hearing loss. Hum. Mutat. 26, 591 (2005).
Tlili, A., Rebeh, I. B., Aifa-Hmani, M., Dhouib, H., Moalla, J., Tlili-Chouchène, J. et al. TMC1 but not TMC2 is responsible for autosomal recessive nonsyndromic hearing impairment in Tunisian families. Audiol. Neurootol. 13, 213–218 (2008).
Saïd, M. B., Hmani-Aifa, M., Amar, I., Baig, S. M., Mustapha, M., Delmaghani, S. et al. High frequency of the p.R34X mutation in the TMC1 gene associated with nonsyndromic hearing loss is due to founder effects. Genet. Test. Mol. Biomarkers 14, 307–311 (2010).
Lui, V. C., Ng, L. J., Sat, E. W. & Cheah, K. S. The human alpha 2(XI) collagen gene (COL11A2): completion of coding information, identification of the promoter sequence, and precise localization within the major histocompatibility complex reveal overlap with the KE5 gene. Genomics 32, 401–412 (1996).
Vikkula, M., Mariman, E. C., Lui, V. C., Zhidkova, N. I., Tiller, G. E., Goldring, M. B. et al. Autosomal dominant and recessive osteochondrodysplasias associated with the COL11A2 locus. Cell 80, 431–437 (1995).
Chen, W., Kahrizi, K., Meyer, N. C., Riazalhosseini, Y., Van Camp, G., Najmabadi, H. et al. Mutation of COL11A2 causes autosomal recessive non-syndromic hearing loss at the DFNB53 locus. J. Med. Genet. 42, e61 (2005).
McGuirt, W. T., Prasad, S. D., Griffith, A. J., Kunst, H. P., Green, G. E., Shpargel, K. B. et al. Mutations in COL11A2 cause nonsyndromic hearing loss (DFNA13). Nat. Genet. 23, 413–419 (1999).
de Leenheer, E. M., Bosman, A. J., Kunst, H. P., Huygen, P. L. & Cremers, C. W. Audiological characteristics of some affected members of a Dutch DFNA13/COL11A2 family. Ann. Otol. Rhinol. Laryngol. 113, 922–929 (2004).
Friedman, T. B., Liang, Y., Weber, J. L., Hinnant, J. T., Barber, T. D., Winata, S. et al. A gene for congenital, recessive deafness DFNB3 maps to the pericentromeric region of chromosome 17. Nat. Genet. 9, 86–91 (1995).
Wang, A., Liang, Y., Fridell, R. A., Probst, F. J., Wilcox, E. R., Touchman, J. W. et al. Association of unconventional myosin MYO15 mutations with human nonsyndromic deafness DFNB3. Science 280, 1447–1451 (1998).
Berg, J. S., Powell, B. C. & Cheney, R. E. A millennial myosin census. Mol. Biol. Cell. 12, 780–794 (2001).
Friedman, T. B., Hinnant, J. T., Ghosh, M., Boger, E. T., Riazuddin, S., Lupski, J. R. et al. DFNB3, spectrum of MYO15A recessive mutant alleles and an emerging genotype-phenotype correlation. Adv. Otorhinolaryngol. 61, 124–130 (2002).
Nal, N., Ahmed, Z. M., Erkal, E., Alper, O. M., Lüleci, G., Dinç, O. et al. Mutational spectrum of MYO15A: the large N-terminal extension of myosin XVA is required for hearing. Hum. Mutat. 28, 1014–1019 (2007).
Kalay, E., Uzumcu, A., Krieger, E., Caylan, R., Uyguner, O., Ulubil-Emiroglu, M. et al. MYO15A (DFNB3) mutations in turkish hearing loss families and functional modeling of a novel motor domain mutation. Am. J. Med. Genet. A 143A, 2382–2389 (2007).
Shearer, A. E., Hildebrand, M. S., Bromhead, C. J., Kahrizi, K., Webster, J. A., Azadeh, B. et al. A novel splice site mutation in the RDX gene causes DFNB24 hearing loss in an Iranian family. Am. J. Med. Genet. A 149A, 555–558 (2009).
Hoeflich, K. P. & Ikura, M. Radixin: cytoskeletal adopter and signaling protein. Int. J. Biochem. Cell Biol. 36, 2131–2136 (2004).
Khan, S. Y., Ahmed, Z. M., Shabbir, M. I., Kitajiri, S., Kalsoom, S., Tasneem, S. et al. Mutations of the RDX gene cause nonsyndromic hearing loss at the DFNB24 locus. Hum. Mutat. 28, 417–423 (2007).
Shearer, A. E., Hildebrand, M. S., Webster, J. A., Kahrizi, K., Meyer, N. C., Jalalvand, K. et al Mutations in the first MyTH4 domain of MYO15A are a common cause of DFNB3 hearing loss. Laryngoscope 119, 727–733 (2009).
He, L. & Hannon, G. J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 (2004).
Hildebrand, M. S., Witmer, P. D., Xu, S., Newton, S. S., Kahrizi, K., Najmabadi, H. et al. miRNA mutations are not a common cause of deafness. Am. J. Med. Genet. A 152A, 646–652 (2010).
Bitner-Glindzicz, M. Hereditary deafness and phenotyping in Humans. Br. Med. Bull. 63, 73–94 (2002).
Choi, S. Y., Kim, Y. E., Ahn, D. B., Kim, T. H., Choi, J. H., Lee, H. R. et al. Construction of a DNA chip for screening of genetic hearing loss. Clin. Exp. Otorhinolaryngol. 2, 44–47 (2009).
Choi, S. Y., Lee, K. Y., Kim, Y. E., Bae, J. W., Oh, S. K., Kim, S. Y. et al. Application of allele-specific primer extension-based microarray for simultaneous multi-gene mutation screening in patients with non-syndromic hearing loss. Int. J. Mol. Med. 25, 315–320 (2010).
Mahdieh, N., Tafsiri, E., Karimipour, M. & Akbari, M. T. Heterozygosity and allele frequencies of the two VNTRs (ApoB and D1S80) in Iranian population. Indian J. Hum. Genet. 11, 31–34 (2005).
Hashemzadeh Chaleshtori, M., Farrokhi, E., Shahrani, M., Kheiri, S., Dolati, M., Hoghooghi Rad, L. et al. High carrier frequency of the GJB2 mutation (35delG) in the north of Iran. Int. J. Pediatr. Otorhinolaryngol. 71, 863–867 (2007).
Mehra, S., Eavey, R. D. & Keamy, D. G. The epidemiology of hearing impairment in the United States: Newborns, children, and adolescents. Otolaryngol. Head Neck Surg. 140, 461–472 (2009).
Hamid, M., Karimipoor, M., Chaleshtori, M. H. & Akbari, M. T. A novel 355-357delGAG mutation and frequency of connexin-26 (GJB2) mutations in Iranian patients. J. Genet. 88, 359–362 (2009).
Sadeghi, A., Sanati, M. H., Alasti, F., Hashemzadeh Chaleshtori, M. & Ataei, M. Mutation analysis of Connexin 26 gene and del in patients with hereditary deafness from two provinces in Iran. Iran. J. Biotechnol. 3, 255–258 (2005).
Sadeghi, A., Sanati, M. H., Alasti, F., Hashemzadeh Chaleshtori, M., Mahmoudian, S. & Ataei, M. Contribution of GJB2 mutations and four common DFNB loci in autosomal recessive non-syndromic hearing impairment in Markazi and Qom provinces of Iran. Iran. J. Biotechnol. 7, 108–111 (2009).
Acknowledgements
We thank Mrs Fatemeh Fardanesh, Mr Mahmoudi, Miss Afrouz Vahdat and staff of the Iran Society of Deaf People Family. This work was supported by the Kawsar Human Research Center and by the Department of Medical Genetics, Faculty of Medicine, Tarbiat Modares University.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mahdieh, N., Rabbani, B., Wiley, S. et al. Genetic causes of nonsyndromic hearing loss in Iran in comparison with other populations. J Hum Genet 55, 639–648 (2010). https://doi.org/10.1038/jhg.2010.96
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2010.96
Keywords
This article is cited by
-
Analysis of deafness susceptibility gene of neonates in northern Guangdong, China
Scientific Reports (2024)
-
Genetic etiology of hearing loss in Iran
Human Genetics (2022)
-
A novel pathogenic variant in the LRTOMT gene causes autosomal recessive non-syndromic hearing loss in an Iranian family
BMC Medical Genetics (2020)
-
Comprehensive genetic testing of Chinese SNHL patients and variants interpretation using ACMG guidelines and ethnically matched normal controls
European Journal of Human Genetics (2020)
-
Whole exome sequencing identifies novel compound heterozygous pathogenic variants in the MYO15A gene leading to autosomal recessive non-syndromic hearing loss
Molecular Biology Reports (2020)