Abstract
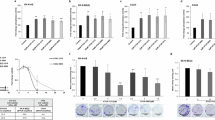
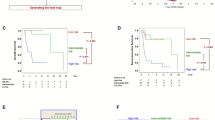
Clinical applications of aberrant DNA methylation to cancer diagnostics and therapeutics are accelerating. Especially, the CpG island methylator phenotype (CIMP), simultaneous methylation of multiple genes, provides information that cannot be obtained by other diagnostic methods and therapeutic opportunities. CIMP is known to be associated with poor or good prognosis depending upon cancer types. We identified that CIMP in neuroblastomas (NBLs) is strongly associated with poor prognosis in Japanese NBL cases (hazard ratio (HR)=22). Almost all NBLs with MYCN amplification displayed CIMP, and even among NBLs without MYCN amplification, NBLs with CIMP had worse prognosis (HR=12). The prognostic power was faithfully reproduced in German NBL cases by the same methods, and also in Italian and Swedish NBL cases with different analytical methods. Mechanistically, methylation silencing of different sets of tumor-suppressor genes is involved in poor prognosis of NBLs with CIMP, and the presence of CIMP is most sensitively detected by methylation of the PCDHB family. For therapeutic purposes, a combination of 5-aza-2′-deoxycytidine, a DNA-demethylating drug, with 13-cis-retinoic acid, a differentiating drug, has been shown to be effective for NBLs in vitro, and further development of a better combination(s) is awaited. Now, epigenetic diagnosis and therapeutics are becoming or have become an important choice for cancer patients.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Church, T. R., Wandell, M., Lofton-Day, C., Mongin, S. J., Burger, M., Payne, S. R. et al. Prospective evaluation of methylated SEPT9 in plasma for detection of asymptomatic colorectal cancer. Gut in press (2013).
Hegi, M. E., Diserens, A. C., Gorlia, T., Hamou, M. F., de Tribolet, N., Weller, M. et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 352, 997–1003 (2005).
Van Neste, L., Herman, J. G., Otto, G., Bigley, J. W., Epstein, J. I. & Van Criekinge, W. The epigenetic promise for prostate cancer diagnosis. Prostate 72, 1248–1261 (2011).
Teodoridis, J. M., Hardie, C. & Brown, R. CpG island methylator phenotype (CIMP) in cancer: causes and implications. Cancer Lett. 268, 177–186 (2008).
Cowan, L. A., Talwar, S., Yang, A. S. & Will, D. N. A. Methylation inhibitors work in solid tumors? A review of the clinical experience with azacitidine and decitabine in solid tumors. Epigenomics 2, 71–86 (2010).
Foulks, J. M., Parnell, K. M., Nix, R. N., Chau, S., Swierczek, K., Saunders, M. et al. Epigenetic drug discovery: targeting DNA methyltransferases. J. Biomol. Screen. 17, 2–17 (2012).
Juergens, R. A., Wrangle, J., Vendetti, F. P., Murphy, S. C., Zhao, M., Coleman, B. et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 1, 598–607 (2011).
Abe, M., Watanabe, N., McDonell, N., Takato, T., Ohira, M., Nakagawara, A. et al. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology 74, 50–60 (2008).
Matei, D., Fang, F., Shen, C., Schilder, J., Arnold, A., Zeng, Y. et al. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 72, 2197–2205 (2012).
Toyota, M., Ahuja, N., Ohe-Toyota, M., Herman, J. G., Baylin, S. B. & Issa, J. P. CpG island methylator phenotype in colorectal cancer. Proc. Natl Acad. Sci. USA 96, 8681–8686 (1999).
Ogino, S., Nosho, K., Kirkner, G. J., Kawasaki, T., Meyerhardt, J. A., Loda, M. et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut 58, 90–96 (2009).
Shen, L., Toyota, M., Kondo, Y., Lin, E., Zhang, L., Guo, Y. et al. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc. Natl Acad. Sci. USA 104, 18654–18659 (2007).
Yagi, K., Akagi, K., Hayashi, H., Nagae, G., Tsuji, S., Isagawa, T. et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin. Cancer Res. 16, 21–33 (2010).
Suzuki, H., Igarashi, S., Nojima, M., Maruyama, R., Yamamoto, E., Kai, M. et al. IGFBP7 is a p53-responsive gene specifically silenced in colorectal cancer with CpG island methylator phenotype. Carcinogenesis 31, 342–349 (2010).
Hinoue, T., Weisenberger, D. J., Pan, F., Campan, M., Kim, M., Young, J. et al. Analysis of the association between CIMP and BRAF in colorectal cancer by DNA methylation profiling. PLoS One 4, e8357 (2009).
Kim, H., Kim, Y. H., Kim, S. E., Kim, N. G. & Noh, S. H. Concerted promoter hypermethylation of hMLH1, p16INK4A, and E-cadherin in gastric carcinomas with microsatellite instability. J. Pathol. 200, 23–31 (2003).
Toyota, M., Ahuja, N., Suzuki, H., Itoh, F., Ohe-Toyota, M., Imai, K. et al. Aberrant methylation in gastric cancer associated with the CpG island methylator phenotype. Cancer Res. 59, 5438–5442 (1999).
Marsit, C. J., Houseman, E. A., Christensen, B. C., Eddy, K., Bueno, R., Sugarbaker, D. J. et al. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer Res. 66, 10621–10629 (2006).
Shen, L., Ahuja, N., Shen, Y., Habib, N. A., Toyota, M., Rashid, A. et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J. Natl Cancer Inst. 94, 755–761 (2002).
Strathdee, G., Appleton, K., Illand, M., Millan, D. W., Sargent, J., Paul, J. et al. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am. J. Pathol. 158, 1121–1127 (2001).
Roman-Gomez, J., Jimenez-Velasco, A., Agirre, X., Castillejo, J. A., Navarro, G., Calasanz, M. J. et al. CpG island methylator phenotype redefines the prognostic effect of t(12;21) in childhood acute lymphoblastic leukemia. Clin. Cancer Res. 12, 4845–4850 (2006).
Maruyama, R., Toyooka, S., Toyooka, K. O., Harada, K., Virmani, A. K., Zochbauer-Muller, S. et al. Aberrant promoter methylation profile of bladder cancer and its relationship to clinicopathological features. Cancer Res. 61, 8659–8663 (2001).
Brock, M. V., Gou, M., Akiyama, Y., Muller, A., Wu, T. T., Montgomery, E. et al. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin. Cancer Res. 9, 2912–2919 (2003).
Wei, S. H., Chen, C. M., Strathdee, G., Harnsomburana, J., Shyu, C. R., Rahmatpanah, F. et al. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin. Cancer Res. 8, 2246–2252 (2002).
Kusano, M., Toyota, M., Suzuki, H., Akino, K., Aoki, F., Fujita, M. et al. Genetic, epigenetic, and clinicopathologic features of gastric carcinomas with the CpG island methylator phenotype and an association with Epstein-Barr virus. Cancer 106, 1467–1479 (2006).
Noushmehr, H., Weisenberger, D. J., Diefes, K., Phillips, H. S., Pujara, K., Berman, B. P. et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 (2010).
Turcan, S., Rohle, D., Goenka, A., Walsh, L. A., Fang, F., Yilmaz, E. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–483 (2012).
Dang, L., White, D. W., Gross, S., Bennett, B. D., Bittinger, M. A., Driggers, E. M. et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 (2009).
Xu, W., Yang, H., Liu, Y., Yang, Y., Wang, P., Kim, S. H. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011).
Figueroa, M. E., Abdel-Wahab, O., Lu, C., Ward, P. S., Patel, J., Shih, A. et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 (2010).
Maris, J. M. Recent advances in neuroblastoma. N. Engl. J. Med. 362, 2202–2211 (2010).
Cohn, S. L., Pearson, A. D., London, W. B., Monclair, T., Ambros, P. F., Brodeur, G. M. et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J. Clin. Oncol. 27, 289–297 (2009).
Jaenisch, R. & Bird, A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33 (Suppl), 245–254 (2003).
Li, E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3, 662–673 (2002).
Abe, M., Ohira, M., Kaneda, A., Yagi, Y., Yamamoto, S., Kitano, Y. et al. CpG island methylator phenotype is a strong determinant of poor prognosis in neuroblastomas. Cancer Res. 65, 828–834 (2005).
Abe, M., Westermann, F., Nakagawara, A., Takato, T., Schwab, M. & Ushijima, T. Marked and independent prognostic significance of the CpG island methylator phenotype in neuroblastomas. Cancer Lett. 247, 253–258 (2007).
Banelli, B., Brigati, C., Di Vinci, A., Casciano, I., Forlani, A., Borzi, L. et al. A pyrosequencing assay for the quantitative methylation analysis of the PCDHB gene cluster, the major factor in neuroblastoma methylator phenotype. Lab. Invest. 92, 458–465 (2011).
Kiss, N. B., Kogner, P., Johnsen, J. I., Martinsson, T., Larsson, C. & Geli, J. Quantitative global and gene-specific promoter methylation in relation to biological properties of neuroblastomas. BMC Med. Genet. 13, 83 (2012).
Schwab, M., Alitalo, K., Klempnauer, K. H., Varmus, H. E., Bishop, J. M., Gilbert, F. et al. Amplified DNA with limited homology to myc cellular oncogene is shared by human neuroblastoma cell lines and a neuroblastoma tumour. Nature 305, 245–248 (1983).
Brodeur, G. M., Seeger, R. C., Schwab, M., Varmus, H. E. & Bishop, J. M. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 224, 1121–1124 (1984).
Seeger, R. C., Brodeur, G. M., Sather, H., Dalton, A., Siegel, S. E., Wong, K. Y. et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N. Engl. J. Med. 313, 1111–1116 (1985).
Grau, E., Martinez, F., Orellana, C., Canete, A., Yanez, Y., Oltra, S. et al. Hypermethylation of apoptotic genes as independent prognostic factor in neuroblastoma disease. Mol. Carcinog. 50, 153–162 (2011).
Alaminos, M., Davalos, V., Ropero, S., Setien, F., Paz, M. F., Herranz, M. et al. EMP3, a myelin-related gene located in the critical 19q13.3 region, is epigenetically silenced and exhibits features of a candidate tumor suppressor in glioma and neuroblastoma. Cancer Res. 65, 2565–2571 (2005).
Alaminos, M., Davalos, V., Cheung, N. K., Gerald, W. L. & Esteller, M. Clustering of gene hypermethylation associated with clinical risk groups in neuroblastoma. J. Natl Cancer Inst. 96, 1208–1219 (2004).
Misawa, A., Inoue, J., Sugino, Y., Hosoi, H., Sugimoto, T., Hosoda, F. et al. Methylation-associated silencing of the nuclear receptor 1I2 gene in advanced-type neuroblastomas, identified by bacterial artificial chromosome array-based methylated CpG island amplification. Cancer Res. 65, 10233–10242 (2005).
Asada, K., Watanabe, N., Nakamura, Y., Ohira, M., Westermann, F., Scwab, M. et al. Stronger prognostic power of the CpG island methylator phenotype than methylation of individual genes in neuroblastomas. Jpn J. Clin. Oncol. in press (2013).
Schulte, J. H., Lim, S., Schramm, A., Friedrichs, N., Koster, J., Versteeg, R. et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 69, 2065–2071 (2009).
Ueda, R., Suzuki, T., Mino, K., Tsumoto, H., Nakagawa, H., Hasegawa, M. et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J. Am. Chem. Soc. 131, 17536–17537 (2009).
George, R. E., Lahti, J. M., Adamson, P. C., Zhu, K., Finkelstein, D., Ingle, A. M. et al. Phase I study of decitabine with doxorubicin and cyclophosphamide in children with neuroblastoma and other solid tumors: a Children's Oncology Group study. Pediatr. Blood Cancer 55, 629–638 (2010).
Weisenberger, D. J., Siegmund, K. D., Campan, M., Young, J., Long, T. I., Faasse, M. A. et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 38, 787–793 (2006).
Acknowledgements
This study was supported by Grants-in-Aid for the Third-Term Cancer Control Strategy Program from the Ministry of Health, Labour and Welfare, Japan, and by the National Cancer Center Research and Development Fund, Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Asada, K., Abe, M. & Ushijima, T. Clinical application of the CpG island methylator phenotype to prognostic diagnosis in neuroblastomas. J Hum Genet 58, 428–433 (2013). https://doi.org/10.1038/jhg.2013.64
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2013.64
Keywords
This article is cited by
-
Epigenetically regulated PCDHB15 impairs aggressiveness of metastatic melanoma cells
Clinical Epigenetics (2022)
-
Functional test of PCDHB11, the most human-specific neuronal surface protein
BMC Evolutionary Biology (2016)