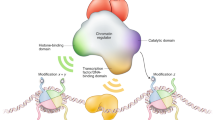
Abstract
Histones function both positively and negatively in the regulation of gene expression, mainly governed by post-translational modifications on specific amino acid residues. Although histone modifications are not necessarily prerequisite codes, they may still serve as good epigenetic indicators of chromatin state associated with gene activation or repression. In particular, six emerging classes of histone H3 modifications are subjected for epigenome profiling by the International Human Epigenome Consortium. In general, transcription start sites of actively transcribed genes are marked by trimethylated H3K4 (H3K4me3) and acetylated H3K27 (H3K27ac), and active enhancers can be identified by enrichments of both monomethylated H3K4 (H3K4me1) and H3K27ac. Gene bodies of actively transcribed genes are associated with trimethylated H3K36 (H3K36me3). Gene repression can be mediated through two distinct mechanisms involving trimethylated H3K9 (H3K9me3) and trimethylated H3K27 (H3K27me3). Enrichments of these histone modifications on specific loci, or in genome wide, in given cells can be analyzed by chromatin immunoprecipitation (ChIP)-based methods using an antibody directed against the site-specific modification. When performing ChIP experiments, one should be careful about the specificity of antibody, as this affects the data interpretation. If cell samples with preserved histone–DNA contacts are available, evaluation of histone modifications, in addition to DNA methylaion, at specific gene loci would be useful for deciphering the epigenome state for human genetics studies.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Luger, K., Dechassa, M. L. & Tremethick, D. J. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell. Biol 13, 436–447 (2012).
Woodcock, C. L., Skoultchi, A. I. & Fan, Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 14, 17–25 (2006).
The ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Kimura, H. Histone dynamics in living cells revealed by photobleaching. DNA Repair (Amst) 4, 939–950 (2005).
Song, C. X., Yi, C. & He, C. Mapping recently identified nucleotide variants in the genome and transcriptome. Nat Biotechnol. 30, 1107–1116 (2012).
Tan, L. & Shi, Y. G. Tet family proteins and 5-hydroxymethylcytosine in development and disease. Development 139, 1895–1902 (2012).
Smith, Z. D. & Meissner, A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 14, 204–220 (2013).
Bannister, A. J. & Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 21, 381–395 (2011).
Greer, E. L. & Shi, Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 13, 343–357 (2012).
Yun, M., Wu, J., Workman, J. L. & Li, B. Readers of histone modifications. Cell Res. 21, 564–578 (2011).
Henikoff, S. & Shilatifard, A. Histone modification: cause or cog? Trends Genet. 27, 389–396 (2011).
Barski, A., Cuddapah, S., Cui, K., Roh, T. Y., Schones, D. E., Wang, Z. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Guenther, M. G., Levine, S. S., Boyer, L. A., Jaenisch, R. & Young, R. A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130, 77–88 (2007).
Malik, S. & Bhaumik, S. R. Mixed lineage leukemia: histone H3 lysine 4 methyltransferases from yeast to human. FEBS J. 277, 1805–1821 (2010).
Blair, L. P., Cao, J., Zou, M. R., Sayegh, J. & Yan, Q. Epigenetic regulation by lysine demethylase 5 (KDM5) enzymes in cancer. Cancers (Basel) 3, 1383–1404 (2011).
Shilatifard, A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu. Rev. Biochem. 81, 65–95 (2012).
Zhang, Y., Chen, A., Yan, X. M. & Huang, G. Disordered epigenetic regulation in MLL-related leukemia. Int. J. Hematol. 96, 428–437 (2012).
Wang, G. G., Song, J., Wang, Z., Dormann, H. L., Casadio, F., Li, H. et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459, 847–851 (2009).
Musselman, C. A., Lalonde, M. E., Cote, J. & Kutateladze, T. G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 19, 1218–1227 (2012).
Ooi, S. K., Qiu, C., Bernstein, E., Li, K., Jia, D., Yang, Z. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007).
Vermeulen, M., Mulder, K. W., Denissov, S., Pijnappel, W. W., van Schaik, F. M., Varier, R. A. et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007).
Bian, C., Xu, C., Ruan, J., Lee, K. K., Burke, T. L., Tempel, W. et al. Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation. EMBO J. 30, 2829–2842 (2011).
Heintzman, N. D., Stuart, R. K., Hon, G., Fu, Y., Ching, C. W., Hawkins, R. D. et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 (2007).
Creyghton, M. P., Cheng, A. W., Welstead, G. G., Kooistra, T., Carey, B. W., Steine, E. J. et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl Acad. Sci. USA 107, 21931–21936 (2010).
Rada-Iglesias, A., Bajpai, R., Swigut, T., Brugmann, S. A., Flynn, R. A. & Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470, 279–283 (2011).
Spicuglia, S. & Vanhille, L. Chromatin signatures of active enhancers. Nucleus 3, 126–131 (2012).
Pekowska, A., Benoukraf, T., Zacarias-Cabeza, J., Belhocine, M., Koch, F., Holota, H. et al. H3K4 tri-methylation provides an epigenetic signature of active enhancers. EMBO J. 30, 4198–4210 (2011).
De Santa, F., Barozzi, I., Mietton, F., Ghisletti, S., Polletti, S., Tusi, B. K. et al. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 8, e1000384 (2010).
Outchkourov, N. S., Muino, J. M., Kaufmann, K., van Ijcken, W. F., Koerkamp, M. J., van Leenen, D. et al. Balancing of histone H3K4 methylation states by the Kdm5c/SMCX histone demethylase modulates promoter and enhancer function. Cell Rep 3, 1071–1079 (2013).
Wagner, E. J. & Carpenter, P. B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell. Biol. 13, 115–126 (2012).
Duns, G., van den Berg, E., van Duivenbode, I., Osinga, J., Hollema, H., Hofstra, R. M. et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 70, 4287–4291 (2010).
Hampsey, M. & Reinberg, D. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113, 429–432 (2003).
Lee, J. S. & Shilatifard, A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat. Res. 618, 130–134 (2007).
Venkatesh, S., Smolle, M., Li, H., Gogol, M. M., Saint, M., Kumar, S. et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 489, 452–455 (2012).
Kim, S., Kim, H., Fong, N., Erickson, B. & Bentley, D. L. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc. Natl Acad. Sci. USA 108, 13564–13569 (2011).
de Almeida, S. F., Grosso, A. R., Koch, F., Fenouil, R., Carvalho, S., Andrade, J. et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 18, 977–983 (2011).
Luco, R. F., Pan, Q., Tominaga, K., Blencowe, B. J., Pereira-Smith, O. M. & Misteli, T. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010).
Pradeepa, M. M., Sutherland, H. G., Ule, J., Grimes, G. R. & Bickmore, W. A. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 8, e1002717 (2012).
Grewal, S. I. & Jia, S. Heterochromatin revisited. Nat. Rev. Genet. 8, 35–46 (2007).
Probst, A. V. & Almouzni, G. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 27, 177–185 (2011).
Klose, R. J., Yamane, K., Bae, Y., Zhang, D., Erdjument-Bromage, H., Tempst, P. et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442, 312–316 (2006).
Whetstine, J. R., Nottke, A., Lan, F., Huarte, M., Smolikov, S., Chen, Z. et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 (2006).
Tahiliani, M., Koh, K. P., Shen, Y., Pastor, W. A., Bandukwala, H., Brudno, Y. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Zee, B. M., Levin, R. S., Xu, B., LeRoy, G., Wingreen, N. S. & Garcia, B. A. In vivo residue-specific histone methylation dynamics. J. Biol. Chem. 285, 3341–3350 (2010).
Rea, S., Eisenhaber, F., O'Carroll, D., Strahl, B. D., Sun, Z. W., Schmid, M. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C. et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107, 323–337 (2001).
Matsui, T., Leung, D., Miyashita, H., Maksakova, I. A., Miyachi, H., Kimura, H. et al. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 (2010).
Maison, C. & Almouzni, G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell. Biol. 5, 296–304 (2004).
Motamedi, M. R., Hong, E. J., Li, X., Gerber, S., Denison, C., Gygi, S. et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol. Cell. 32, 778–790 (2008).
Nozawa, R. S., Nagao, K., Masuda, H. T., Iwasaki, O., Hirota, T., Nozaki, N. et al. Human POGZ modulates dissociation of HP1alpha from mitotic chromosome arms through Aurora B activation. Nat. Cell Biol. 12, 719–727 (2010).
Canzio, D., Liao, M., Naber, N., Pate, E., Larson, A., Wu, S. et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature 496, 377–381 (2013).
Nozawa, R. S., Nagao, K., Igami, K. T., Shibata, S., Shirai, N., Nozaki, N. et al. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat. Struct. Mol. Biol. 20, 566–573 (2013).
Almouzni, G. & Probst, A. V. Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus 2, 332–338 (2011).
Morey, L. & Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 35, 323–332 (2010).
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011).
Wang, H., Wang, L., Erdjument-Bromage, H., Vidal, M., Tempst, P., Jones, R. S. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004).
Stock, J. K., Giadrossi, S., Casanova, M., Brookes, E., Vidal, M., Koseki, H. et al. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 9, 1428–1435 (2007).
Mosammaparast, N. & Shi, Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179 (2010).
Albert, M. & Helin, K. Histone methyltransferases in cancer. Semin. Cell Dev. Biol. 21, 209–220 (2010).
Pedersen, M. T. & Helin, K. Histone demethylases in development and disease. Trends Cell Biol. 20, 662–671 (2010).
Nestorov, P., Tardat, M. & Peters, A. H. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr. Top. Dev. Biol. 104, 243–291 (2013).
Brockdorff, N. Noncoding RNA and Polycomb recruitment. RNA 19, 429–442 (2013).
Kassis, J. A. & Brown, J. L. Polycomb group response elements in Drosophila and vertebrates. Adv. Genet. 81, 83–118 (2013).
Chadwick, B. P. & Willard, H. F. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc. Natl Acad. Sci. USA 101, 17450–17455 (2004).
Bernstein, B. E., Mikkelsen, T. S., Xie, X., Kamal, M., Huebert, D. J., Cuff, J. et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 (2006).
Vastenhouw, N. L. & Schier, A. F. Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 24, 374–386 (2012).
Ballare, C., Lange, M., Lapinaite, A., Martin, G. M., Morey, L., Pascual, G. et al. Phf19 links methylated Lys36 of histone H3 to regulation of Polycomb activity. Nat. Struct. Mol. Biol. 19, 1257–1265 (2012).
Brien, G. L., Gambero, G., O'Connell, D. J., Jerman, E., Turner, S. A., Egan, C. M. et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 19, 1273–1281 (2012).
Britton, L. M., Gonzales-Cope, M., Zee, B. M. & Garcia, B. A. Breaking the histone code with quantitative mass spectrometry. Expert Rev. Proteomics. 8, 631–643 (2011).
Sidoli, S., Cheng, L. & Jensen, O. N. Proteomics in chromatin biology and epigenetics: elucidation of post-translational modifications of histone proteins by mass spectrometry. J. Proteomics 75, 3419–3433 (2012).
Collas, P. The current state of chromatin immunoprecipitation. Mol. Biotechnol. 45, 87–100 (2010).
Dahl, J. A., Reiner, A. H. & Collas, P. Fast genomic muChIP-chip from 1,000 cells. Genome Biol. 10, R13 (2009).
Adli, M. & Bernstein, B. E. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat. Protoc. 6, 1656–1668 (2011).
Zhou, V. W., Goren, A. & Bernstein, B. E. Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 (2011).
Zhang, Y., Liu, T., Meyer, C. A., Eeckhoute, J., Johnson, D. S., Bernstein, B. E. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Egelhofer, T. A., Minoda, A., Klugman, S., Lee, K., Kolasinska-Zwierz, P., Alekseyenko, A. A. et al. An assessment of histone-modification antibody quality. Nat. Struct. Mol. Biol. 18, 91–93 (2011).
Clayton, A. L., Hazzalin, C. A. & Mahadevan, L. C. Enhanced histone acetylation and transcription: a dynamic perspective. Mol. Cell 23, 289–296 (2006).
Kimura, H., Hayashi-Takanaka, Y., Goto, Y., Takizawa, N. & Nozaki, N. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell. Struct. Funct. 33, 61–73 (2008).
Hayashi-Takanaka, Y., Yamagata, K., Wakayama, T., Stasevich, T. J., Kainuma, T., Tsurimoto, T. et al. Tracking epigenetic histone modifications in single cells using Fab-based live endogenous modification labeling. Nucleic Acids Res. 39, 6475–6488 (2011).
Thurman, R. E., Rynes, E., Humbert, R., Vierstra, J., Maurano, M. T., Haugen, E. et al. The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012).
Feng, S., Jacobsen, S. E. & Reik, W. Epigenetic reprogramming in plant and animal development. Science 330, 622–627 (2010).
Mikeska, T., Bock, C., Do, H. & Dobrovic, A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Rev. Mol. Diagn. 12, 473–487 (2012).
Gomez, D., Shankman, L. S., Nguyen, A. T. & Owens, G. K. Detection of histone modifications at specific gene loci in single cells in histological sections. Nat. Methods 10, 171–177 (2013).
Portela, A. & Esteller, M. Epigenetic modifications and human disease. Nat. Biotechnol. 28, 1057–1068 (2010).
Rodriguez-Paredes, M. & Esteller, M. Cancer epigenetics reaches mainstream oncology. Nat. Med. 17, 330–339 (2011).
Butler, J. S., Koutelou, E., Schibler, A. C. & Dent, S. Y. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics 4, 163–177 (2012).
You, J. S. & Jones, P. A. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 22, 9–20 (2012).
Acknowledgements
I thank Yuko Sato for preparing the figures, Timothy J Stasevich for proofreading and Yoko Hayashi-Takanaka for providing experimental data. Their comments on the manuscript have also been greatly helpful. This work was supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Japan Science and Technology Agency, Core Research for Evolutional Science and Technology, and New Energy and Industrial Technology Development Organization.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kimura, H. Histone modifications for human epigenome analysis. J Hum Genet 58, 439–445 (2013). https://doi.org/10.1038/jhg.2013.66
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2013.66
Keywords
This article is cited by
-
Sex-specific epigenetics drive low GPER expression in gastrointestinal smooth muscles in type 2 diabetic mice
Scientific Reports (2024)
-
Epigenetic repression of CHCHD2 enhances survival from single cell dissociation through attenuated Rho A kinase activity
Cellular and Molecular Life Sciences (2024)
-
ENPP1 inhibits the transcription activity of the hepatitis B virus pregenomic promoter by upregulating the acetylation of LMNB1
Archives of Virology (2024)
-
Molecular characterization and modulated expression of histone acetyltransferases during cold response of the tick Dermacentor silvarum (Acari: Ixodidae)
Parasites & Vectors (2023)
-
Crosstalk of hepatocyte nuclear factor 4a and glucocorticoid receptor in the regulation of lipid metabolism in mice fed a high-fat-high-sugar diet
Lipids in Health and Disease (2022)