Abstract
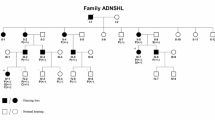
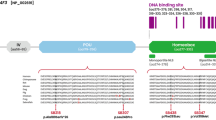
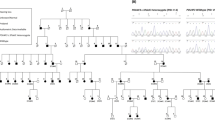
Hearing impairment, or deafness (in its most severe form), is one of the most common human sensory disorders. There have been several reports of autosomal dominant mutations in the POU4F3 gene, which is associated with non-syndromic hearing loss. In this study, we identified a novel heterozygous mutation (c.602delT, p.L201fs) in the gene POU4F3 by taking advantage of whole-exome sequencing, which was validated by Sanger sequencing and completely co-segregated within a large hearing impaired Chinese family. We have focused on this pedigree since 2002, and we have mapped a deafness locus named DFNA42 (which has been renamed DFNA52, OMIM entry 607683) via a genome-wide scan. Furthermore, we analyzed this mutational variant and found that it was located at the beginning of the first functional domain of POU4F3, which could theoretically impair the function of POU4F3. We have identified a novel frameshift mutation in the POU4F3 gene. Further functional studies of variants of this specific gene are needed to illustrate the pathogenic mechanism(s) that underlie hearing impairment.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Morton, N. E. Genetic epidemiology of hearing impairment. Ann. NY Acad. Sci. 630, 16–31 (1991).
Kim, S. Y., Kim, A. R., Kim, N. K., Kim, M. Y., Jeon, E. H., Kim, B. J. et al. Strong founder effect of p.P240L in CDH23 in Koreans and its significant contribution to severe-to-profound nonsyndromic hearing loss in a Korean pediatric population. J. Transl. Med. 13, 263 (2015).
Xia, J., Deng, H., Feng, Y., Zhang, H., Pan, Q., Dai, H. et al. A novel locus for autosomal dominant nonsyndromic hearing loss identified at 5q31.1-32 in a Chinese pedigree. J. Hum. Genet. 47, 635–640 (2002).
Venkatesh, M. D., Moorchung, N. & Puri, B. Genetics of non syndromic hearing loss. Med. J. Armed Forces India 71, 363–368 (2015).
Ku, C. S., Naidoo, N. & Pawitan, Y. Revisiting Mendelian disorders through exome sequencing. Hum. Genet. 129, 351–370 (2011).
Fukui, H., Wong, H. T., Beyer, L. A., Case, B. G., Swiderski, D. L., Di Polo, A. et al. BDNF gene therapy induces auditory nerve survival and fiber sprouting in deaf Pou4f3 mutant mice. Sci. Rep. 2, 838 (2012).
Baek, J. I., Oh, S. K., Kim, D. B., Choi, S. Y., Kim, U. K., Lee, K. Y. et al. Targeted massive parallel sequencing: the effective detection of novel causative mutations associated with hearing loss in small families. Orphanet J. Rare. Dis. 7, 60 (2012).
Frydman, M., Vreugde, S., Nageris, B. I., Weiss, S., Vahava, O. & Avraham, K. B. Clinical characterization of genetic hearing loss caused by a mutation in the POU4F3 transcription factor. Arch. Otolaryngol. Head Neck Surg. 126, 633–637 (2000).
Nolan, L. S., Jagutpal, S. S., Cadge, B. A., Woo, P. & Dawson, S. J. Identification and functional analysis of common sequence variants in the DFNA15 gene, Brn-3c. Gene 400, 89–97 (2007).
Xiang, M., Gan, L., Li, D., Chen, Z. Y., Zhou, L., O'Malley, B. W. Jr. et al. Essential role of POU-domain factor Brn-3c in auditory and vestibular hair cell development. Proc. Natl Acad. Sci. USA 94, 9445–9450 (1997).
Collin, R. W., Chellappa, R., Pauw, R. J., Vriend, G., Oostrik, J., van Drunen, W. et al. Missense mutations in POU4F3 cause autosomal dominant hearing impairment DFNA15 and affect subcellular localization and DNA binding. Hum. Mutat. 29, 545–554 (2008).
Lee, H. K., Park, H. J., Lee, K. Y., Park, R. & Kim, U. K. A novel frameshift mutation of POU4F3 gene associated with autosomal dominant non-syndromic hearing loss. Biochem. Biophys. Res. Commun. 396, 626–630 (2010).
Weiss, S., Gottfried, I., Mayrose, I., Khare, S. L., Xiang, M., Dawson, S. J. et al. The DFNA15 deafness mutation affects POU4F3 protein stability, localization, and transcriptional activity. Mol. Cell. Biol. 23, 7957–7964 (2003).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Vahava, O., Morell, R., Lynch, E. D., Weiss, S., Kagan, M. E., Ahituv, N. et al. Mutation in transcription factor POU4F3 associated with inherited progressive hearing loss in humans. Science 279, 1950–1954 (1998).
van Drunen, F. J., Pauw, R. J., Collin, R. W., Kremer, H., Huygen, P. L. & Cremers, C. W. Vestibular impairment in a Dutch DFNA15 family with an L289F mutation in POU4F3. Audiol. Neurootol. 14, 303–307 (2009).
Acknowledgements
We greatly thank the pedigree members for agreeing to participate in this study. We also appreciate the help and advice of our colleagues. This study was supported by the National Basic Research Program of China (also called 973 Program) (2012CB517902; 2014CB541702) and the National Nature Science Foundation of China (81300833; 31301023).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Cai, X., Li, Y., Xia, L. et al. Exome sequencing identifies POU4F3 as the causative gene for a large Chinese family with non-syndromic hearing loss. J Hum Genet 62, 317–320 (2017). https://doi.org/10.1038/jhg.2016.102
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2016.102
This article is cited by
-
LINE-1 global DNA methylation, iron homeostasis genes, sex and age in sudden sensorineural hearing loss (SSNHL)
Human Genomics (2023)
-
Ramifications of POU4F3 variants associated with autosomal dominant hearing loss in various molecular aspects
Scientific Reports (2023)
-
Highly variable hearing loss due to POU4F3 (c.37del) is revealed by longitudinal, frequency specific analyses
European Journal of Human Genetics (2023)
-
Coffin-Siris syndrome in two chinese patients with novel pathogenic variants of ARID1A and SMARCA4
Genes & Genomics (2022)
-
Gene therapy development in hearing research in China
Gene Therapy (2020)