Abstract
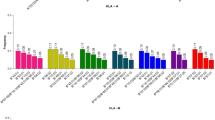
The south of Tunisia is characterized by marked ethnic diversity, highlighted by the coexistence of native Berbers with Blacks, Jews and Arab-speaking populations. Despite this heterogeneity, genetic anthropology studies investigating the origin of current Southern Tunisians were rarely reported. We examined human leukocyte antigen (HLA) class I (A, B) and class II (DRB1, DQB1) gene profiles of 250 unrelated Southern Tunisians, and compared them with those of Arab-speaking communities, along with Mediterranean and sub-Sahara African populations using genetic distances, neighbor-joining dendrograms, correspondence and haplotype analysis. In total, 137 HLA alleles were detected, which comprised 32 HLA-A, 52 HLA-B, 32 DRB1 and 21 DQB1 alleles. The most frequent alleles were HLA-A*02:01(18.02%), HLA-B*50:01 (9.11%), HLA-DRB1*07:01 (22.06%) and HLA-DQB1*02:01 (17.21%). All pairs of HLA loci show significant linkage disequilibrium. The four loci depict negative Fnd (the normalized deviate of the homozygosity) values indicating an overall trend to balancing selection. Southern Tunisians appear to be closely related to others Tunisian populations including Berbers, North Africans and Iberians. On the contrary, Southern Tunisians were distinct from Palestinian, Lebanese and Jordanian Middle Eastern Arab-speaking population, despite the deep Arab incursions and Arabization that affected Southern Tunisia. In addition, Southern Tunisians were distant from many sub-Saharan communities, evidenced by genetic distance analysis. Collectively, this indicates a limited genetic contribution of Arab invasion and Black caravans on the makeup of Southern Tunisian gene pool.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Shiina, T., Hosomichi, K., Inoko, H. & Kulski, J. K. The HLA genomic loci map: expression, interaction, diversity, and disease. J. Hum. Genet. 54, 15–39 (2009).
Robinson, J., Soormally, A. R., Hayhurst, J. D. & Marsh, S. G. The IPD-IMGT/HLA database—new developments in reporting HLA variation. Hum. Immunol. 77, 233–237 (2016).
Vina, M. A., Hollenbach, J. A., Lyke, K. E., Sztein, M. B., Maiers, M., Klitz, W. et al. Tracking human migrations by the analysis of the distribution of HLA alleles, lineages and haplotypes in closed and open populations. Philos. Trans. R. Soc. Lond. Ser. B 367, 820–829 (2012).
Brett, M. & Fentress, E. The Berbers, (Blackwell Publishers, Oxford, UK, 1997).
Stearns, P. N. & Leonard, W. L. in The Encyclopedia of World History: Ancient, Medieval, and Modern, Chronologically Arranged 6th edn, 129–131 (Houghton Mifflin Harcourt, New York, NY, USA, 2001).
Austin, R. A. The Transaharian Slave Trade. Essays in the Economic History of the Atlantic Slave Trade, (New York Academy Press, New York, NY, 1979).
Lucette, V. & Abraham, L. U. Juifs en terre d’islam: les communautés de Djerba 13 (éd. Archives contemporaines, Paris, 1991).
Ibn Khaldūn, A. in The Muqaddimah: An Introduction to History (Trans. Franz Rosenthal, ed. Dawood, N. J., 1967) (abridged).
Hajjej, A., Hajjej, G., Almawi, W. Y., Kaabi, H., El-Gaaied, A. & Hmida, S. HLA class I and class II polymorphism in a population from south-eastern Tunisia (Gabes Area). Int. J. Immunogenet. 38, 191–199 (2011).
Hajjej, A., Hmida, S., Kaabi, H., Dridi, A., Jridi, A., El Gaaled, A. et al. HLA genes in Southern Tunisians (Ghannouch area) and their relationship with other Mediterraneans. Eur. J. Med. Genet. 49, 43–56 (2006).
Miller, S. A., Dykes, D. D. & Polesky, H. F. A sample salting out procedure for extraction DNA from human nucleated cells. Nucleic Acids Res. 16, 1215–1218 (1988).
Buyse, I., Decorte, R., Baens, M., Cuppens, H., Semana, G., Emonds, M. P. et al. Rapid DNA typing of class II HLA antigens using the polymerase chain reaction and reverse dot blot hybridization. Tissue Antigens 41, 1–14 (1993).
Marsh, S. G. E., Albert, E. D., Bodmer, W. F., Bontrop, R. E., Dupont, B., Erlich, H. A et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens 75, 291–455 (2010).
Excoffier, L. & Slatkin, M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 12, 921–927 (1995).
Schneider, S., Kueffer, J. M., Roessli, D. & Excoffier, L. Arlequin: A Software Environment for the Analysis of Population Genetics Data, (Genetics and Biometry Lab, Geneva, Switzerland, 1996).
Imanishi, T., Akaza, T., Kimura, A., Tokunaga, K., Gojrobi, T. in HLA 1991 Vol. I (eds Tsuji, K., Aizawa, M. & Sasazuki, T.) 76–79 (Oxford University Press, Oxford, UK, 1992).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Nei, M. Genetic distances between populations. Am. Nat. 106, 283 (1972).
Nei, M., Tajima, Y. & Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J. Mol. Evol. 19, 153–170 (1983).
Young, F. W. & Bann, C. M. in Statistical Computing Environments for Social Researches (eds Stine, R. A. & Fox, J.) 207–236 (Sage Publications, New York, NY, 1996).
Lancaster, A. K., Nelson, M. P., Single, R. M., Meyer, D. & Thomson, G. in Pacific Symposium on Biocomputing Vol. 8 (eds Altman, R. B., Dunker, K., Hunter, L., Jung, T. & Klein, T.) 514–525 (World Scientific, Singapore, Singapore, 2003).
Lancaster, A. K., Single, R. M., Solberg, O. D., Nelson, M. P. & Thomson, G. PyPop update—a software pipeline for large-scale multilocus population genomics. Tissue Antigens 69, 192–197 (2007).
Ewens, W. The sampling theory of selectively neutral alleles. Theor. Pop. Biol. 3, 87–112 (1972).
Watterson, G. The homozygosity test of neutrality. Genetics 88, 405–417 (1978).
Slatkin, M. An exact test for neutrality based on the Ewens sampling distribution. Genet. Res. 64, 71–74 (1994).
Slatkin, M. A correction to the exact test based on the Ewens sampling distribution. Genet. Res. 68, 259–260 (1996).
Glantz, S. A. Primer of Biostatistics 7th edn (McGraw-Hill, New York, NY, 2012).
Gomez-Casado, E., del Moral, P., Martinez-Laso, J., García-Gómez, A., Allende, L., Silvera-Redondo et al. HLA gene in Arabic-Speaking Moroccans: close relatedness to Berbers and Iberians. Tissue Antigens 55, 239–249 (2000).
Comas, D., Mateu, E., Calafell, F., Pérez-Lezaun, A., Bosch, E., Martínez-Arias, R. et al. HLA class I and class II DNA typing and the origin of Basques. Tissue Antigens 51, 30–40 (1998).
Clayton, J. & Lonjou, C. in Genetic Diversity of HLA. Functional and Medical Implications Vol. 1 (ed. Charron, D.) 665–820 (EDK: Paris, 1997).
Arnaiz-Villena, A., Benmamar, D., Alvarez, M., Diaz-Campos, N., Varela, P., Gomez-Casado, E. et al. HLA allele and haplotype frequencies in Algerians. Relatedness to Spaniards and Basques. Hum. Immunol. 43, 259–268 (1995).
Hajjej, A., Kâabi, H., Sellami, M. H., Dridi, A., Jeridi, A., El Borgi, W. et al. The contribution of HLA class I and II alleles and haplotypes to the investigation of the evolutionary history of Tunisians. Tissue Antigens 68, 153–162 (2006).
Hajjej, A., Sellami, M. H., Kaabi, Hajjej, G., El-Gaaied, A., Boukef, K. et al. HLA class I and class II polymorphisms in Tunisian Berbers. Ann. Hum. Biol. 38, 156–164 (2011).
Hajjej, A., Almawi, W. Y., Hattab, L., El-Gaaied, A. & Hmida, S. HLA class I and class II alleles and haplotypes confirm the Berber Origin of the Present Day Tunisian Population. PLoS ONE 10, e0136909 (2015).
Sánchez-Velasco, P., Karadsheh, N. S., García-Martín, A., Ruíz de Alegría, C. & Leyva-Cobián, F. Molecular analysis of HLA allelic frequencies and haplotypes in Jordanians and comparison with other related populations. Hum. Immunol. 62, 901–909 (2001).
Galgani, A., Mancino, G., Martínez-Labarga, C., Cicconi, R., Mattei, M., Amicosante, M. et al. HLA-A, -B and -DRB1 allele frequencies in Cyrenaica population (Libya) and genetic relationships with other populations. Hum. Immunol. 74, 52–59 (2013).
Arnaiz-Villena, A., Elaiwa, N., Silvera, C., Rostom, A., Moscoso, J., Gómez-Casado, E. et al The origin of Palestinians and their genetic relatedness with other Mediterranean populations [retraction] Hum. Immunol. 62, 889–900 (2001).
Hmida, S., Gauthier, Dridi, A., Quillivic, F., Genetet, B., Boukef et al. HLA class II gene polymorphism in Tunisians. Tissue Antigens 45, 63–68 (1995).
Izaabel, H., Garchon, H. J., Caillat-Zucman, S., Beaurain, G., Akhayat, O., Bach, J. F. et al. HLA class II DNA polymorphism in a Moroccan population from the Souss, Agadir area. Tissue Antigens 51, 106–110 (1998).
Canossi, A., Piancatelli, D., Aureli, A., Oumhani, K., Ozzella, G., Del Beato, T. et al. Correlation between genetic HLA class I and II polymorphisms and anthropological aspects in the Chaouya population from Morocco (Arabic speaking). Tissue Antigens 76, 177–193 (2010).
Brick, C., Atouf, O., Bouayad, A . & Essakalli, M. Moroccan study of HLA (-A, -B, -C, -DR, -DQ) polymorphism in 647 unrelated controls: Updating data. Mol. Cell. Probes 29, 197–207 (2015).
Martinez-Laso, J., De Juan, Martinez-Quiles, N., Gomez-Casado, E., Cuadrado, E. & Arnaiz-Villena, A. The contribution of the HLA-A, -B, -C and -DR, -DQ DNA typing to the study of the origins of Spaniards and Basques. Tissue Antigens 45, 237–245 (1995).
Roitberg-Tambur, A., Witt, C. S., Friedmann, Safirman, C., Sherman, Battat, S. et al. Comparative analysis of HLA polymorphism at the serologic and molecular level in Moroccan and Ashkenazi Jews. Tissue Antigens 46, 104–110 (1995).
Arnaiz-Villena, A., Karin, M., Bendikuze, N., Gomez-Casado, E., Moscoso, J., Silvera, C. et al. HLA alleles and haplotypes in the Turkish population: relatedness to Kurds, Armenians and other Mediterraneans. Tissue Antigens 57, 308–317 (2001).
Imanishi, T., Akaza, T., Kimura, A., Tokunaga, K. & Gjobori, T. in HLA 1991 Vol. 1 (eds Tsuji, K., Aizawa, M. & Sasazuki, T.) 1065–1220 (Oxford University Press, Oxford, UK, 1992).
Sanchez-Velasco, P., Gomez-Casado, E., Martinez-Laso, J., Moscoso, J., Zamora, J., Lowy, E. et al. HLA alleles in isolated populations from North Spain: origin of the Basques and the ancient Iberians. Tissue Antigens 61, 384–392 (2003).
Single, R., Meyer, D., Mack, S. J., Lancaster, A., Nelson, M. P. & Erlich, H. et al. in Immunobiology of the Human MHC (ed. Jansen, J. A.) 705–746 (International Histocompatibility Working Group Press, Seattle, WA, 2006).
Mack, S. J., Tu, B., Lazaro, A., Yang, R., Lancaster, A. K., Cao, K. et al. HLA-A, -B, -C, and -DRB1 allele and haplotype frequencies distinguish Eastern European Americans from the general European American population. Tissue Antigens 73, 17–32 (2009).
Satta, Y., O’hUigin, C., Takahata, N. & Klein, J. Intensity of natural selection at the major histocompatibility complex loci. Proc. Natl Acad. Sci. USA 91, 7184–7188 (1994).
Abdennaji Guenounou, B., Yacoubi Loueslati, B., Buhler, S., Hmida, S., Ennafaa, H., Khodjet-Elkhil, H. et al. HLA class II genetic diversity in Southern Tunisia and the Mediterranean area. Int. J. Immunogenet. 33, 93–103 (2006).
Fadhlaoui-Zid, K., Buhler, S., Dridi, A., Benammar El Gaaied, A. & Sanchez-Mazas, A. Polymorphism of HLA class II genes in Berbers from Southern Tunisia. Tissue Antigens 76, 416–420 (2010).
Mahfoudh, N., Ayadi, I., Kamoun, A., Ammar, R., Mallek, B. & Maalej, L. et al. Analysis of HLA-A, -B, -C, -DR, -DQ polymorphisms in the South Tunisian population and a comparison with other populations. Ann Hum Biol. 40, 41–47 (2013).
Ayed, K., Ayed-Jendoubi, S., Sfar, I., Labonne, M. P. & Gebuhrer, L. HLA class-I and HLA class-II phenotypic, gene and haplotypic frequencies in Tunisians by using molecular typing data. Tissue Antigens 64, 520–532 (2004).
Solberg, O. D., Mack, S. J., Lancaster, A. K., Single, R. M., Tsai, Y. & Sanchez-Mazas, A. et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum. Immunol. 69, 43–464 (2008).
Apanius, V., Penn, O. & Slev, P. R. et al. The nature of selection on the major histocompatibility complex. Crit. Rev. Immunol. 15, 179–224 (1997).
Arnaiz-Villena, A., Gomez-Casado, E. & Martinez-Laso, J. Population genetic relationships between Mediterranean populations determined by HLA allele distribution and a historic perspective. Tissue Antigens 60, 111–121 (2002).
Julien, C. A. Histoire de l’Afrique du Nord (Masson et Cie, Paris, 1953).
Murdok, G. P. Africa, Its Peoples and Their Cultural History (McGrawHill, New York, NY, 1959).
Arnaiz-Villena, A., Iliakis, P., González-Hevilla, M., Longás, J., Gómez-Casado, E., Sfyridaki, K. et al. The origin of Cretan populations as determined by characterization of HLA alleles. Tissue Antigens 53, 213–226 (1999).
Fischer, T. in The International Geography (ed. Mill, H. R.) 368–377 (Appleton and Company, New York, NY and London, UK, 1920).
Stallaert, C. Ethnogenesis and Ethnicity in Spain: A Historical–Anthropological Approach to Casticismo (Proyecto A: Barcelona, Spain, 1998).
Botigué, L. M., Henn, B. M., Gravel, S., Maples, B. K., Gignoux, C. R. & Corona, E. et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc. Natl Acad. Sci. USA 110, 11791–11796 (2013).
Currat, M., Poloni, E. S. & Sanchez-Mazas, A. Human genetic differentiation across the Strait of Gibraltar. Evol. Biol. 10, 237 (2010).
Le Bon, G. Arab Civilisation (Libairie Firmin Diderot, France, 1884).
Mark, A. & Tessler, A. History of the Israeli-Palestinian Conflict Vol. 329 (Indiana University Press, Bloomington, IN, 1994).
Arnaiz-Villena, A., Dimitroski, K., Pacho, A., Moscoso, J., Gómez-Casado, E. & Silvera-Redondo, C. et al. HLA genes in Macedonians and the Sub-Saharan origin of the Greeks. Tissue Antigens 57, 118–127 (2001).
Dork, T., El-Harith, E. -H. A., Stuhrmann, M., Macek, M. Jr, Egan, M., Cutting, G. R. et al. Evidence for a common ethnic origin of cystic fibrosis mutation 3120+1G-to-A in diverse populations. Am. J. Hum. Genet. 63, 656–662 (1998).
Herodotus. History (Gredos, Madrid, Spain, 1989).
Petlichkovski, A., Efinska-Mladenovska, O., Trajkov, D., Arsov, T., Strezova, A. & Spiroski, M. High-resolution typing of HLA-DRB1 locus in the Macedonian population. Tissue Antigens 64, 486–491 (2004).
Fadhlaoui-Zid, K., Martinez-Cruz, B., Khodjet-el-khil, H., Mendizabal, I., Benammar-Elgaaied, A. & Comas, D. Genetic structure of Tunisian ethnic groups revealed by paternal lineages. Am. J. Phys. Anthropol. 146, 271–280 (2011).
Fadhlaoui-Zid, K., Plaza, S., Calafell, F., Ben Amor, M., Comas, D. & Bennamar El gaaied, A. Mitochondrial DNA heterogeneity in Tunisian Berbers. Ann. Hum. Genet. 68, 222–233 (2004).
Frigi, S., Cherni, L., Fadhlaoui-Zid, K. & Benammar-Elgaaied, A. Ancient local evolution of African mtDNA haplogroups in Tunisian Berber populations. Hum. Biol. 82, 367–384 (2010).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Hajjej, A., Almawi, W., Hattab, L. et al. The investigation of the origin of Southern Tunisians using HLA genes. J Hum Genet 62, 419–429 (2017). https://doi.org/10.1038/jhg.2016.146
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2016.146
This article is cited by
-
Contribution of HLA class I (A, B, C) and HLA class II (DRB1, DQA1, DQB1) alleles and haplotypes in exploring ethnic origin of central Tunisians
BMC Medical Genomics (2024)
-
HLA-A, -B, -C, -DRB1 and -DQB1 allele and haplotype frequencies in Lebanese and their relatedness to neighboring and distant populations
BMC Genomics (2022)