Abstract
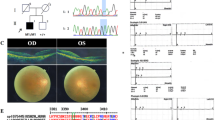
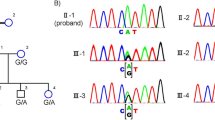
Inherited retinal degeneration (IRD) are a group of genetically heterogeneous disease of which retinitis pigmentosa (RP) and Leber congenital amaurosis (LCA) are the most common and severe type. In our study we had taken three unrelated South Indian consanguineous IRD families. Homozygosity mapping was done using Affymetrix 250K Nsp1 GeneChip in each of LCA, Cone-Rod dystrophy (CRD) and autosomal recessive RP (arRP) families followed by targeted re-sequencing by next generation sequencing (NGS) on Illumina MiSeq. Known candidate genes ranging from 1–8 in numbers within the homozygous blocks were identified by homozygosity mapping and targeted NGS revealed the causative mutations; RDH12 c.832A>C, ABCA4 c.1462G>T, CDHR1c.1384_1392delCTCCTGGACinsG, in the LCA, CRD and arRP families, respectively. The identified mutations were validated by Sanger sequencing, segregation in the families and their absence in 200 control chromosomes. Homozygosity mapping guided targeted NGS, especially when more numbers of known candidate genes within the homozygous blocks are observed is a comprehensive method for mutation identification. Molecular data from a larger retinal degenerative disease cohort would reveal the spectrum and prevalence of mutations and genes in Indian population. Molecular diagnosis also aids in genetic counseling, offering carrier and prenatal testing to family members.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Shanks, M. E., Downes, S. M., Copley, R. R., Lise, S., Broxholme, J., Hudspith, K. A. et al. Next-generation sequencing (NGS) as a diagnostic tool for retinal degeneration reveals a much higher detection rate in early-onset disease. Eur. J. Hum. Genet. 21, 274–280 (2013).
Jin, X., Qu, L. H., Meng, X. H., Xu, H. W. & Yin, Z. Q. Detecting genetic variations in hereditary retinal dystrophies with next-generation sequencing technology. Mol. Vis. 20, 553–560 (2014).
Rivolta, C., Sharon, D., DeAngelis, M. M. & Dryja, T. P. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum. Mol. Genet. 11, 1219–1227 (2002).
Fahim, A. T., Daiger, S. P. & Weleber, R. G. in Retinitis Pigmentosa Overview (eds Pagon, R. A., Adam, M. P., Ardinger, H. H., Wallace, S. E., Amemiya, A., Bean, L. J. H. et al.) (Seattle, WA, USA, 1993).
Dharmaraj, S. R., Silva, E. R., Pina, A. L., Li, Y. Y., Yang, J. M., Carter, C. R. et al. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic Genet. 21, 135–150 (2000).
Chacon-Camacho, O. F. & Zenteno, J. C. Review and update on the molecular basis of Leber congenital amaurosis. World J. Clin. Cases 3, 112–124 (2015).
Gu, S. M., Thompson, D. A., Srikumari, C. R., Lorenz, B., Finckh, U., Nicoletti, A. et al. Mutations in RPE65 cause autosomal recessive childhood-onset severe retinal dystrophy. Nat. Genet. 17, 194–197 (1997).
den Hollander, A. I., Lopez, I., Yzer, S., Zonneveld, M. N., Janssen, I. M., Strom, T. M. et al. Identification of novel mutations in patients with Leber congenital amaurosis and juvenile RP by genome-wide homozygosity mapping with SNP microarrays. Invest. Ophthalmol. Vis. Sci. 48, 5690–5698 (2007).
den Hollander, A. I., Roepman, R., Koenekoop, R. K. & Cremers, F. P. Leber congenital amaurosis: genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 27, 391–419 (2008).
Koenekoop, R. K. The gene for Stargardt disease, ABCA4, is a major retinal gene: a mini-review. Ophthalmic Genet. 24, 75–80 (2003).
Mullins, R. F., Kuehn, M. H., Radu, R. A., Enriquez, G. S., East, J. S., Schindler, E. I. et al. Autosomal recessive retinitis pigmentosa due to ABCA4 mutations: clinical, pathologic, and molecular characterization. Invest. Ophthalmol. Vis. Sci. 53, 1883–1894 (2012).
Valverde, D., Riveiro-Alvarez, R., Aguirre-Lamban, J., Baiget, M., Carballo, M., Antinolo, G. et al. Spectrum of the ABCA4 gene mutations implicated in severe retinopathies in Spanish patients. Invest. Ophthalmol. Vis. Sci. 48, 985–990 (2007).
Kannabiran, C., Singh, H., Sahini, N., Jalali, S. & Mohan, G. Mutations in TULP1, NR2E3, and MFRP genes in Indian families with autosomal recessive retinitis pigmentosa. Mol. Vis. 18, 1165–1174 (2012).
Ramprasad, V. L., Soumittra, N., Nancarrow, D., Sen, P., McKibbin, M., Williams, G. A. et al. Identification of a novel splice-site mutation in the Lebercilin (LCA5) gene causing Leber congenital amaurosis. Mol. Vis. 14, 481–486 (2008).
Glockle, N., Kohl, S., Mohr, J., Scheurenbrand, T., Sprecher, A., Weisschuh, N. et al. Panel-based next generation sequencing as a reliable and efficient technique to detect mutations in unselected patients with retinal dystrophies. Eur. J. Hum. Genet. 22, 99–104 (2014).
Srilekha, S., Arokiasamy, T., Srikrupa, N. N., Umashankar, V., Meenakshi, S., Sen, P. et al. Homozygosity Mapping in Leber Congenital Amaurosis and Autosomal Recessive Retinitis Pigmentosa in South Indian Families. PLoS ONE 10, e0131679 (2015).
Mannan, A. U., Singh, J., Lakshmikeshava, R., Thota, N., Singh, S., Sowmya, T. S. et al. Detection of high frequency of mutations in a breast andor ovarian cancer cohort: implications of embracing a multi-gene panel in molecular diagnosis in India. J. Hum. Genet. 61, 515–522 (2016).
Adzhubei, I. A., Schmidt, S., Peshkin, L., Ramensky, V. E., Gerasimova, A., Bork, P. et al. A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 4, 1073–1081 (2009).
Schwarz, J. M., Cooper, D. N., Schuelke, M. & Seelow, D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods 11, 361–362 (2014).
Reva, B., Antipin, Y. & Sander, C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 39, e118 (2011).
Ferrer-Costa, C., Gelpi, J. L., Zamakola, L., Parraga, I., de la Cruz, X. & Orozco, M. PMUT: a web-based tool for the annotation of pathological mutations on proteins. Bioinformatics 21, 3176–3178 (2005).
Li, B., Krishnan, V. G., Mort, M. E., Xin, F., Kamati, K. K., Cooper, D. N. et al. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25, 2744–2750 (2009).
Woods, C. G., Cox, J., Springell, K., Hampshire, D. J., Mohamed, M. D., McKibbin, M. et al. Quantification of homozygosity in consanguineous individuals with autosomal recessive disease. Am. J. Hum. Genet. 78, 889–896 (2006).
Kirin, M., McQuillan, R., Franklin, C. S., Campbell, H., McKeigue, P. M. & Wilson, J. F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE 5, e13996 (2010).
Hildebrandt, F., Heeringa, S. F., Ruschendorf, F., Attanasio, M., Nurnberg, G., Becker, C. et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 5, e1000353 (2009).
Thompson, D. A., Janecke, A. R., Lange, J., Feathers, K. L., Hubner, C. A., McHenry, C. L. et al. Retinal degeneration associated with RDH12 mutations results from decreased 11-cis retinal synthesis due to disruption of the visual cycle. Hum. Mol. Genet. 14, 3865–3875 (2005).
Valverde, D., Pereiro, I., Vallespin, E., Ayuso, C., Borrego, S. & Baiget, M. Complexity of phenotype-genotype correlations in Spanish patients with RDH12 mutations. Invest. Ophthalmol. Vis. Sci. 50, 1065–1068 (2009).
Maugeri, A., Klevering, B. J., Rohrschneider, K., Blankenagel, A., Brunner, H. G., Deutman, A. F. et al. Mutations in the ABCA4 (ABCR) gene are the major cause of autosomal recessive cone-rod dystrophy. Am. J. Hum. Genet. 67, 960–966 (2000).
Cideciyan, A. V., Swider, M., Aleman, T. S., Tsybovsky, Y., Schwartz, S. B., Windsor, E. A. et al. ABCA4 disease progression and a proposed strategy for gene therapy. Hum. Mol. Genet. 18, 931–941 (2009).
Nikopoulos, K., Avila-Fernandez, A., Corton, M., Lopez-Molina, M. I., Perez-Carro, R., Bontadelli, L. et al. Identification of two novel mutations in CDHR1 in consanguineous Spanish families with autosomal recessive retinal dystrophy. Sci. Rep. 5, 13902 (2015).
Acknowledgements
We thank Indian Council of Medical Research (ICMR), Govt. of India for the grant 54/1/2010-BMS and SRF Fellowship – N0.45/2/2014-HUM-BMS, and all the patients and their family for their kind cooperation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Sundaramurthy, S., Swaminathan, M., Sen, P. et al. Homozygosity mapping guided next generation sequencing to identify the causative genetic variation in inherited retinal degenerative diseases. J Hum Genet 61, 951–958 (2016). https://doi.org/10.1038/jhg.2016.83
Received:
Revised:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/jhg.2016.83