Abstract
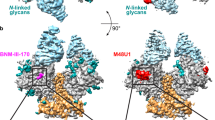
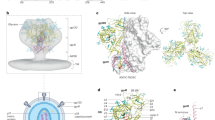
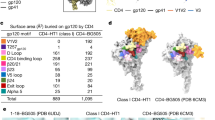
The remarkable diversity, glycosylation and conformational flexibility of the human immunodeficiency virus type 1 (HIV-1) envelope (Env), including substantial rearrangement of the gp120 glycoprotein upon binding the CD4 receptor, allow it to evade antibody-mediated neutralization. Despite this complexity, the HIV-1 Env must retain conserved determinants that mediate CD4 binding. To evaluate how these determinants might provide opportunities for antibody recognition, we created variants of gp120 stabilized in the CD4-bound state, assessed binding of CD4 and of receptor-binding-site antibodies, and determined the structure at 2.3 Å resolution of the broadly neutralizing antibody b12 in complex with gp120. b12 binds to a conformationally invariant surface that overlaps a distinct subset of the CD4-binding site. This surface is involved in the metastable attachment of CD4, before the gp120 rearrangement required for stable engagement. A site of vulnerability, related to a functional requirement for efficient association with CD4, can therefore be targeted by antibody to neutralize HIV-1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Korber, B. et al. Timing the ancestor of the HIV-1 pandemic strain. Science 288, 1789–1796 (2000)
Joint United National Programme on HIV/AIDS. 2006 Report on the global AIDS epidemic. 〈http://www.aids.org/en/HIVdata/2006GlobalReport/〉 (2006)
Weiss, R. A. et al. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature 316, 69–72 (1985)
Wyatt, R. & Sodroski, J. The HIV-1 envelope glycoproteins: fusogens, antigens and immunogens. Science 280, 1884–1888 (1998)
Parren, P. W., Moore, J. P., Burton, D. R. & Sattentua, Q. J. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13 (suppl. A) S137–S162 (1999)
Kowalski, M. L. et al. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237, 1351–1355 (1987)
Lu, M., Blackow, S. & Kim, P. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nature Struct. Biol. 2, 1075–1082 (1995)
Starcich, B. R. et al. Identification and characterization of conserved and variable regions of the envelope gene HTLV-III/LAV, the retrovirus of AIDS. Cell 45, 637–648 (1986)
Wyatt, R. et al. The antigenic structure of the human immunodeficiency virus gp120 envelope glycoprotein. Nature 393, 705–711 (1998)
Kwong, P. D. et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682 (2002)
Wei, X. et al. Antibody neutralization and escape by HIV-1. Nature 422, 307–312 (2003)
Burton, D. R. Antibodies, viruses and vaccines. Nature Rev. Immunol. 2, 706–713 (2002)
Luftig, M. A. et al. Structural basis for HIV-1 neutralization by a gp41 fusion intermediate-directed antibody. Nature Struct. Mol. Biol. 13, 740–747 (2006)
Dalgleish, A. G. et al. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312, 763–767 (1984)
Feng, F., Broder, C. C., Kennedy, P. E. & Berger, E. A. HIV-1 entry co-factor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272, 872–877 (1996)
Chan, D. C., Fass, D., Berger, J. M. & Kim, P. S. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997)
Weissenhorn, W., Dessen, A., Harrison, S. C., Skehel, J. J. & Wiley, D. C. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 (1997)
Kwong, P. D. et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659 (1998)
Thali, M. et al. Characterization of conserved human immunodeficiency virus type 1 (HIV-1) gp120 neutralization epitopes exposed upon gp120–CD4 binding. J. Virol. 67, 3978–3988 (1993)
Chen, B. et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433, 834–841 (2005)
Zhang, M. Y. et al. Improved breadth and potency of an HIV-1-neutralizing single-chain antibody by random mutagenesis and sequential antigen panning. J. Mol. Biol. 335, 209–219 (2004)
Rits-Volloch, S., Frey, G., Harrison, S. C. & Chen, B. Restraining the conformation of HIV-1 gp120 by removing a flexible loop. EMBO J. 25, 5026–5035 (2006)
Profy, A. T. et al. Epitopes recognized by the neutralizing antibodies of an HIV-1-infected individual. J. Immunol. 144, 4641–4647 (1990)
Burton, D. R. & Montefiori, D. C. The antibody response in HIV-1 infection. AIDS 11, (suppl. A)587–598 (1997)
Burton, D. R. et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994)
Trkola, A. et al. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4 IgG. J. Virol. 69, 6609–6617 (1995)
Trkola, A. et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70, 1100–1108 (1996)
Parren, P. W. et al. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75, 8340–8347 (2001)
Saphire, E. O. et al. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293, 1155–1159 (2001)
Sundberg, E. J. & Mariuzza, R. A. Molecular recognition in antibody-antigen complexes. Adv. Protein Chem. 61, 119–160 (2002)
Connolly, M. L. The molecular surface package. J. Mol. Graph. 11, 139–141 (1993)
Pantophlet, R. et al. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77, 642–658 (2003)
Zwick, M. B. et al. Molecular features of the broadly neutralizing immunoglobulin G1 b12 required for recognition of human immunodeficiency virus type 1 gp120. J. Virol. 77, 5863–5876 (2003)
Yang, X. et al. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 78, 12975–12986 (2004)
Arthos, J. et al. Biochemical and biological characterization of a dodecameric CD4-Ig fusion protein. Implications for therapeutic and vaccine strategies. J. Biol. Chem. 277, 11456–11464 (2002)
Li, Y. et al. Neutralizing specificity mapping in complex polyclonal sera (AIDS Vaccine 2006, Amsterdam, 2006)
Rossmann, M. G. et al. Structure of a human common cold virus and functional relationship to other picornoviruses. Nature 317, 145–153 (1985)
Wiley, D. C., Wilson, I. A. & Skehel, J. J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289, 373–378 (1981)
Butters, T. D. et al. Effects of N-butyldeoxynojirimycin and the Lec3.2.8.1 mutant phenotype on N-glycan processing in Chinese hamster ovary cells: application to glycoprotein crystallization. Protein Sci. 8, 1696–1701 (1999)
Barouch, D. H. et al. A human T-cell leukemia virus type 1 regulatory element enhances the immunogenicity of human immunodeficiency virus type 1 DNA vaccines in mice and nonhuman primates. J. Virol. 79, 8828–8834 (2005)
Kwong, P. D. et al. Probability analysis of variational crystallization and its application to gp120, the exterior envelope glycoprotein of type 1 human immunodeficiency virus (HIV-1). J. Biol. Chem. 274, 4115–4123 (1999)
Weik, M. et al. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc. Natl Acad. Sci. USA 97, 623–628 (2000)
Lusty, C. J. A gentle vapor diffusion technique for cross-linking of protein crystals for cryocrystallography. J. Appl. Crystallogr. 32, 106–112 (1999)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Navaza, J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A 50, 157–163 (1994)
McRee, D. E. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125, 156–165 (1999)
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Prabakaran, P. et al. Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J. Biol. Chem. 281, 15829–15836 (2006)
Fleury, D., Daniels, R. S., Skehel, J. J., Knossow, M. & Bizebard, T. Structural evidence for recognition of a single epitope by two distinct antibodies. Proteins 40, 572–578 (2000)
Acknowledgements
We thank B. Graham, D. Hamer, S. Harrison, R. Pantophlet, W. Schief, L. Shapiro, I. Wilson and members of the Structural Biology Section, VRC, for discussions and comments on the manuscript; M. Gao for assistance with PDB deposit; H. Katinger for antibody 2G12; G. Lin for suggesting the use of swainsonine; S. Majeed for preparation of Fab 17b; J. Nelson for assistance with b12 ELISAs; M. Posner for antibody F105; J. Robinson for antibodies 17b, 1.5e and F91; J. Stuckey for assistance with figures; M. Venturi for assistance with gp120 production methodology; and the NIH AIDS Research and Reference Reagent Program for CD4. Support for this work was provided by the Intramural Research Program of the NIH, by the International AIDS Vaccine Initiative, by a grant from the Bill and Melinda Gates Foundation Grand Challenges in Global Heath Initiative, and by grants from the NIH. Use of SER-CAT at the Advanced Photon Source was supported by the US Department of Energy, Basic Energy Sciences, Office of Science.
Author Contributions T.Z. and P.D.K. carried out structure-based stabilization, SPR analyses and structural determinations; L.X. and G.J.N. constructed gp120 substitutions and developed and implemented a high-throughput gp120-production system suitable for crystallization; B.D. and R.W. carried out ITC characterizations; A.J.H., M.B.Z. and D.R.B. provided b12, b3, b6, b11 and b13, and mutant b12 binding; D.V.R. and J.A. provided D1D2-Igαtp and associated SPR analyses; S.-H.X., X.Y. and J.S. provided OD1 and preliminary design and antigenic analyses; and M.-Y.Z. and D.S.D. provided m6, m14 and m18. All authors contributed to the manuscript preparation.
Coordinates and structure factors have been deposited in the Protein Data Bank and may be obtained from the authors (accession codes 2nxy–2ny6 for the nine variant gp120 molecules with CD4 and 17b; accession code 2ny7 for the b12–gp120 complex)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Coordinates and structure factors have been deposited in the Protein Data Bank and may be obtained from the authors (accession codes 2nxy–2ny6 for the nine variant gp120 molecules with CD4 and 17b; accession code 2ny7 for the b12–gp120 complex). Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1- 7 and Supplementary Figures 1-6 with Legends. (PDF 1638 kb)
Rights and permissions
About this article
Cite this article
Zhou, T., Xu, L., Dey, B. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737 (2007). https://doi.org/10.1038/nature05580
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/nature05580
This article is cited by
-
On the irrationality of rational design of an HIV vaccine in light of protein intrinsic disorder
Archives of Virology (2021)
-
Analysis of the SARS-CoV-2 spike protein glycan shield reveals implications for immune recognition
Scientific Reports (2020)
-
Broadly neutralizing antibodies and vaccine design against HIV-1 infection
Frontiers of Medicine (2020)
-
Antibody responses to viral infections: a structural perspective across three different enveloped viruses
Nature Microbiology (2019)
-
Differential processing of HIV envelope glycans on the virus and soluble recombinant trimer
Nature Communications (2018)