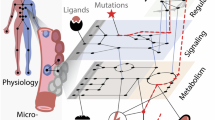
Abstract
Cells employ highly dynamic signaling networks to drive biological decision processes. Perturbations to these signaling networks may attract cells to new malignant signaling and phenotypic states, termed cancer network attractors, that result in cancer development. As different cancer cells reach these malignant states by accumulating different molecular alterations, uncovering these mechanisms represents a grand challenge in cancer biology. Addressing this challenge will require new systems-based strategies that capture the intrinsic properties of cancer signaling networks and provide deeper understanding of the processes by which genetic lesions perturb these networks and lead to disease phenotypes. Network biology will help circumvent fundamental obstacles in cancer treatment, such as drug resistance and metastasis, empowering personalized and tumor-specific cancer therapies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Nash, J.F. Equilibrium points in N-person games. Proc. Natl. Acad. Sci. USA 36, 48–49 (1950).
Nash, J.F. Non-cooperative games. Ann. Math. 54, 286–295 (1951).
Hanahan, D. & Weinberg, R.A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Hanahan, D. & Weinberg, R.A. The hallmarks of cancer. Cell 100, 57–70 (2000).
Davies, H. et al. Mutations of the BRAF gene in human cancer. Nature 417, 949–954 (2002).
Stratton, M.R., Campbell, P.J. & Futreal, P.A. The cancer genome. Nature 458, 719–724 (2009).
Ding, L. et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 481, 506–510 (2012).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 366, 883–892 (2012).
Hou, Y. et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell 148, 873–885 (2012).
Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011).
Wu, M., Pastor-Pareja, J.C. & Xu, T. Interaction between RasV12 and scribbled clones induces tumour growth and invasion. Nature 463, 545–548 (2010).
Ng, P.C. & Henikoff, S. Predicting the effects of amino acid substitutions on protein function. Annu. Rev. Genomics Hum. Genet. 7, 61–80 (2006).
Dixit, A. et al. Sequence and structure signatures of cancer mutation hotspots in protein kinases. PLoS ONE 4, e7485 (2009).
Pazos, F. & Bang, J.-W. Computational prediction of functionally important regions in proteins. Curr. Bioinform. 1, 15–23 (2006).
Fowler, D.M. et al. High-resolution mapping of protein sequence-function relationships. Nat. Methods 7, 741–746 (2010).
Jensen, L.J. et al. Ab initio prediction of human orphan protein function from post-translational modifications and localization features. J. Mol. Biol. 319, 1257–1265 (2002).
Socolich, M. et al. Evolutionary information for specifying a protein fold. Nature 437, 512–518 (2005).
Russ, W., Lowery, D., Mishra, P., Yaffe, M. & Ranganathan, R. Natural-like function in artificial WW domains. Nature 437, 579–583 (2005).
Puntervoll, P. et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31, 3625–3630 (2003).
Lim, W.A. & Pawson, T. Phosphotyrosine signaling: evolving a new cellular communication system. Cell 142, 661–667 (2010).
Seet, B.T., Dikic, I., Zhou, M.M. & Pawson, T. Reading protein modifications with interaction domains. Nat. Rev. Mol. Cell Biol. 7, 473–483 (2006).
Halabi, N., Rivoire, O., Leibler, S. & Ranganathan, R. Protein sectors: Evolutionary units of three-dimensional structure. Cell 138, 774–786 (2009).
Reynolds, K.A., McLaughlin, R. & Ranganathan, R. Hot spots for allosteric regulation on protein surfaces. Cell 147, 1564–1575 (2011).
Wan, P.T. et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 116, 855–867 (2004).
Janes, K.A. et al. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science 310, 1646–1653 (2005).
Lei, K. & Davis, R.J. JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. Proc. Natl. Acad. Sci. USA 100, 2432–2437 (2003).
Lamb, J.A. et al. JunD mediates survival signaling by the JNK signal transduction pathway. Mol. Cell 11, 1479–1489 (2003).
Abreu-Martin, M.T. et al. Fas activates the JNK pathway in human colonic epithelial cells: lack of a direct role in apoptosis. Am. J. Physiol. 276, G599 (1999).
Jeong, H., Mason, S.P., Barabasi, A.L. & Oltvai, Z.N. Lethality and centrality in protein networks. Nature 411, 41–42 (2001).
Shah, S.P. et al. The clonal and mutational evolution spectrum of primary triplenegative breast cancers. Nature advance online publication, doi:10.1038/nature10933 (4 April 2012).
Kauffman, S. & Levin, S. Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 128, 11–45 (1987).
Kauffman, S.A. & Weinberger, E.D. The NK model of rugged fitness landscapes and its application to maturation of the immune response. J. Theor. Biol. 141, 211–245 (1989).
Uribesalgo, I., Benitah, S.A. & Di Croce, L. From oncogene to tumor suppressor: The dual role of Myc in leukemia. Cell Cycle 11, 1757–1764 (2012).
Yang, L., Han, Y., Saurez Saiz, F. & Minden, M.D. A tumor suppressor and oncogene: the WT1 story. Leukemia 21, 868–876 (2007).
Ellis, M.J. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature 486, 353–360 (2012).
Curtis, C. et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Kan, Z. et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010).
Greenman, C. et al. Pattern of somatic mutation in human cancer genomes. Nature 446, 153–158 (2007).
Waddington, C.H. The Strategy of the Genes: a Discussion of Some Aspects of Theoretical Biology (Allen & Unwin, 1957).
Huang, S. & Ingber, D.E. Shape-dependent control of cell growth, differentiation, and apoptosis: switching between attractors in cell regulatory networks. Exp. Cell Res. 261, 91–103 (2000).
Luo, J., Solimini, N.L. & Elledge, S.J. Principles of cancer therapy: oncogene and non-oncogene addiction. Cell 136, 823–837 (2009).
Songyang, Z. et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signaling. Nature 373, 536–539 (1995).
Zhong, Q. et al. Edgetic perturbation models of human inherited disorders. Mol. Syst. Biol. 5, 321 (2009).
Dreze, M. et al. 'Edgetic' perturbation of a C. elegans BCL2 ortholog. Nat. Methods 6, 843–849 (2009).
Pe'er, D. & Hacohen, N. Principles and strategies for developing network models in cancer. Cell 144, 864–873 (2011).
Vidal, M., Cusick, M.E. & Barabási, A.-L.L. Interactome networks and human disease. Cell 144, 986–998 (2011).
Schoeberl, B. et al. Therapeutically targeting ErbB3: a key node in ligand-induced activation of the ErbB receptor-PI3K axis. Sci. Signal. 2, ra31 (2009).
Huang, P.H. et al. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc. Natl. Acad. Sci. USA 104, 12867–12872 (2007).
Miller, M.L.L. et al. Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 1, ra2+ (2008).
Linding, R. et al. Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 (2007).
Mok, J. et al. Deciphering protein kinase specificity through large-scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3, ra12 (2010).
Brinkworth, R.I., Breinl, R.A. & Kobe, B. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc. Natl. Acad. Sci. USA 100, 74–79 (2003).
Turk, B.E. Understanding and exploiting substrate recognition by protein kinases. Curr. Opin. Chem. Biol. 12, 4–10 (2008).
Skerker, J.M. et al. Rewiring the specificity of two-component signal transduction systems. Cell 133, 1043–1054 (2008).
Capra, E.J., Perchuk, B.S., Skerker, J.M. & Laub, M.T. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell 150, 222–232 (2012).
Zarrinpar, A., Park, S.H. & Lim, W.A. Optimization of specificity in a cellular protein interaction network by negative selection. Nature 426, 676–680 (2003).
Wang, X. et al. Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat. Biotechnol. 30, 159–164 (2012).
Brehme, M. et al. Charting the molecular network of the drug target Bcr-Abl. Proc. Natl. Acad. Sci. USA 106, 7414–7419 (2009).
Wong, K.M.M., Hudson, T.J. & McPherson, J.D. Unraveling the genetics of cancer: genome sequencing and beyond. Annu. Rev. Genomics Hum. Genet. 12, 407–430 (2011).
Ledford, H. Big science: the cancer genome challenge. Nature 464, 972–974 (2010).
Bensimon, A., Heck, A.J.R. & Aebersold, R. Mass spectrometry-based proteomics and network biology. Annu. Rev. Biochem. 81, 379–405 (2012).
Geiger, T., Cox, J., Ostasiewicz, P., Wisniewski, J.R. & Mann, M. Super-SILAC mix for quantitative proteomics of human tumor tissue. Nat. Methods 7, 383–385 (2010).
Prill, R.J. et al. Towards a rigorous assessment of systems biology models: the DREAM3 challenges. PLoS ONE 5, e9202 (2010).
Meyer, P. et al. Verification of systems biology research in the age of collaborative competition. Nat. Biotechnol. 29, 811–815 (2011).
Jørgensen, C. et al. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science 326, 1502–1509 (2009).
Pawson, T. & Linding, R. Network medicine. FEBS Lett. 582, 1266–1270 (2008).
Chandarlapaty, S. et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19, 58–71 (2011).
Lee, M.J. et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149, 780–794 (2012).
Erler, J.T. & Linding, R. Network medicine strikes a blow against breast cancer. Cell 149, 731–733 (2012).
Navin, N. et al. Tumor evolution inferred by single-cell sequencing. Nature 472, 90–94 (2011).
Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell 149, 994–1007 (2012).
Pedersen, M.W. et al. Sym004: a novel synergistic anti-epidermal growth factor receptor antibody mixture with superior anticancer efficacy. Cancer Res. 70, 588–597 (2010).
Bendall, S.C. & Nolan, G.P. From single cells to deep phenotypes in cancer. Nat. Biotechnol. 30, 639–647 (2012).
Roque, F.S. et al. Using electronic patient records to discover disease correlations and stratify patient cohorts. PLOS Comput. Biol. 7, e1002141 (2011).
Jensen, P.B., Jensen, L.J. & Brunak, S. Mining electronic health records: towards better research applications and clinical care. Nat. Rev. Genet. 13, 395–405 (2012).
Blumenthal, R.D. & Goldenberg, D.M. Methods and goals for the use of in vitro and in vivo chemosensitivity testing. Mol. Biotechnol. 35, 185–197 (2007).
Hoffman, R.M. Orthotopic mouse models expressing fluorescent proteins for cancer drug discovery. Expert Opin. Drug Discov. 5, 851–866 (2010).
Gonzalez-Angulo, A.M., Hennessy, B.T. & Mills, G.B. Future of personalized medicine in oncology: a systems biology approach. J. Clin. Oncol. 28, 2777–2783 (2010).
Hunter, K.W. Mouse models of cancer: does the strain matter? Nat. Rev. Cancer 12, 144–149 (2012).
Cox, T.R. & Erler, J.T. Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis. Model. Mech. 4, 165–178 (2011).
WHO. World health organization fact sheet 297 (2012). http://www.who.int/mediacentre/factsheets/fs297/en/
Acknowledgements
We apologize to our colleagues whose work could not be cited due to space limitations. We thank all members of the C-SIG (DTU), the ErlerLab (BRIC), M. Yaffe (MIT) and N. Brunner (KU) for critical input on this manuscript. R.L. is a Lundbeck Foundation Fellow and is supported by a Sapere Aude Starting Grant from The Danish Council for Independent Research and a Career Development Award from Human Frontier Science Program. J.T.E. is supported by a Hallas Møller Stipend from the Novo Nordisk Foundation. Visit http://www.networkbio.org/, http://www.lindinglab.org/ and http://www.erlerlab.org/ for more information on cancer-related network biology.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Creixell, P., Schoof, E., Erler, J. et al. Navigating cancer network attractors for tumor-specific therapy. Nat Biotechnol 30, 842–848 (2012). https://doi.org/10.1038/nbt.2345
Published:
Issue date:
DOI: https://doi.org/10.1038/nbt.2345
This article is cited by
-
Identification of anticancer drug target genes using an outside competitive dynamics model on cancer signaling networks
Scientific Reports (2021)
-
WASABI: a dynamic iterative framework for gene regulatory network inference
BMC Bioinformatics (2019)
-
Minimal intervening control of biomolecular networks leading to a desired cellular state
Scientific Reports (2019)
-
A bioprinted human-glioblastoma-on-a-chip for the identification of patient-specific responses to chemoradiotherapy
Nature Biomedical Engineering (2019)
-
Systems Pharmacogenomic Landscape of Drug Similarities from LINCS data: Drug Association Networks
Scientific Reports (2019)