Abstract
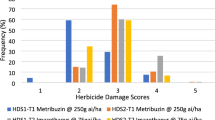
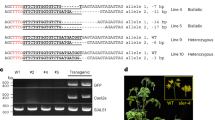
Maize plants resistant to imidazolinone herbicides were engineered through targeted modification of endogenous genes using chimeric RNA/DNA oligonucleotides. A precise single-point mutation was introduced into genes encoding acetohydroxyacid synthase (AHAS), at a position known to confer imidazolinone resistance. Phenotypically normal plants from the converted events (C0) were regenerated from resistant calli and grown to maturity. Herbicide leaf painting confirmed the resistance phenotype in C0 plants and demonstrated the anticipated segregation pattern in C1 progeny. DNA cloning and sequencing of the targeted region in resistant calli and derived C0 and C1 plants confirmed the expected mutation. These results demonstrate that oligonucleotide-mediated gene manipulation can be applied to crop improvement. This approach does not involve genomic integration of transgenes. Since the new trait is obtained through modifying a gene within its normal chromosomal context, position effects, transgene silencing, or other concerns that arise as part of developing transgenic events are avoided.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Shaner, D.L., Anderson, P.C. & Stidham, M.A. Imidazolinones, potent inhibitors of acetohydroxyacid synthase. Plant Physiol. 76, 545–546 (1984).
Stidham, M.A. & Singh, B.K. Imidazolinone–acetohydroxyacid synthase interactions. In The imidazolinone herbicides. (eds Shaner, D. & O'Connor, S.) 71–90 (CRC Press, Boca Raton, FL; 1991).
Sathasivan, K., Haughn, G.W. & Murai, N. Molecular basis of imidazolinone herbicide resistance in Arabidopsis thaliana var. Columbia. Plant Physiol. 97, 1044–1050 (1991).
Ott, K.H., Kwagh, J.G., Stockton, G.W., Sidorov, V. & Kakefuda., G. Rational molecular design and genetic engineering of herbicide-resistant crops by structure modeling and site-directed mutagenesis of acetohydroxyacid synthase. J. Mol. Biol. 263, 359–368 (1996).
Zhu, T. et al. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc. Natl. Acad. Sci. USA 96, 8768–8773 (1999).
Matzke, A.J. Position effects and epigenic silencing of plant transgenes. Curr. Opin. Plant Biol. 1, 142–148 (1998).
Vaucheret, H. Transgene-induced gene silencing in plants. Plant J. 16, 651–659 (1998).
Yoon, K., Cole-Strauss, A. & Kmiec, E.B. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA·DNA oligonucleotide . Proc. Natl. Acad. Sci. USA 93, 2071–2076 (1996).
Cole-Strauss, A. et al. Correction of the mutation responsible for sickle cell anemia by an RNA/DNA oligonucleotide. Science 273, 1386–1389 (1996).
Kren, B.T., Cole-Strauss, A., Kmiec, E.B. & Steer, C.J. Targeted nucleotide exchange in the alkaline phosphatase gene of HuH-7 cells mediated by a chimeric RNA/DNA oligonucleotide. Hepatology 25, 1462–1468 (1997).
Kren, B.T., Bandyopadhyay, P. & Steer, C.J. In vivo site-directed mutagenesis of the Factor IX gene by chimeric RNA/DNA oligonucleotides. Nat. Med. 4B, 285–290 (1998).
Xiang, Y., Cole-Strauss, A., Yoon, K., Gryn, J. & Kmiec, E.B. Targeted gene conversion in a mammalian CD34+-enriched cell population using a chimeric RNA/DNA oligonucleotide. J. Mol. Med. 75, 829–835 (1997).
Alexeev, V. & Yoon, K. Stable and inheritable changes in genotype and phenotype of albino melanocytes induced by an RNA–DNA oligonucleotide. Nat. Biotechnol. 16, 1343–1346 (1998).
Beetham, P.R., Kipp, P.B., Sawycky, X.L., Arntzen, C.J. & May G.D. A tool for functional plant genomics; chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc. Natl. Acad. Sci. USA 96, 8774–8778 (1999).
Cole-Strauss, A., Nöe, A. & Kmiec, E.B. Recombinational repair of genetic mutations. Antisense Nucleic Acid Drug Dev. 7, 211–216 (1997).
Fang, L.Y., Cross, P.R., Chen, C.-H. & Lillis, M. Sequence of two acetohydroxyacid synthase genes from Zea mays. Plant Mol. Biol. 18, 1185–1187 (1992).
Armstrong, C.L, Green, C.E. & Phillips, R.L. Development and availability germplasm with high type II culture formation response. Maize Genet. Coop. Newslett. 65, 92–93 (1991).
Kirihara, J.A. Selection of stable transformants from Black Mexican Sweet maize suspension cultures. In The maize handbook (eds Freeling, M. & Walbot, V.) 663–670 (Springer, New York, NY; 1994).
Ohl, S., Offringa, R., van den Elzen, P.J.M. & Hooykaas, P.J.J. Gene replacement in plants. In Homologous recombination and gene silencing in plants. (ed. Paszkowski, J.) 191–217 (Kluwer Academic Publishers, Dordrecht, The Netherlands; 1994).
Paszkowski. J., Baur, M., Bogucki, A. & Potrykus, I. Gene targeting in plants. EMBO J. 7, 4021–4026 (1988).
Puchta, H. Towards targeted transformation in plants. Trends Plant Sci. 3, 77–78 (1998).
Bergelson, J., Purrington, C.B. & Wichmann, G. Promiscuity in transgenic plants. Nature 393, 25 (1998).
Register, J.C. et al. Structure and function of selectable and non-selectable transgenes in maize after introduction by particle bombardment. Plant Mol. Biol. 25, 951–961 (1994).
Acknowledgements
We thank Donglong Liu and Susan Jayne for advice on leaf painting and herbicide treatment, Marjorie Rudert, Toni Lockwood, Jacob Simmons, Michelle Siegrist, William Van Zante, the Pioneer Hi-Bred media prep and greenhouse staff for excellent technical assistance. We also thank Wesley Bruce, Susan Jayne, and Roger Kemble for reviewing this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, T., Mettenburg, K., Peterson, D. et al. Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat Biotechnol 18, 555–558 (2000). https://doi.org/10.1038/75435
Received:
Accepted:
Issue date:
DOI: https://doi.org/10.1038/75435
This article is cited by
-
Synergistic mutations of two rapeseed AHAS genes confer high resistance to sulfonylurea herbicides for weed control
Theoretical and Applied Genetics (2020)
-
Relaxed chromatin induced by histone deacetylase inhibitors improves the oligonucleotide-directed gene editing in plant cells
Journal of Plant Research (2018)
-
Current and future editing reagent delivery systems for plant genome editing
Science China Life Sciences (2017)
-
Analyses of point mutation repair and allelic heterogeneity generated by CRISPR/Cas9 and single-stranded DNA oligonucleotides
Scientific Reports (2016)
-
Homology-based double-strand break-induced genome engineering in plants
Plant Cell Reports (2016)